Transcription Factor NsdD Regulates the Expression of Genes Involved in Plant Biomass-Degrading Enzymes, Conidiation, and Pigment Biosynthesis in Penicillium oxalicum
- PMID: 29980558
- PMCID: PMC6121985
- DOI: 10.1128/AEM.01039-18
Transcription Factor NsdD Regulates the Expression of Genes Involved in Plant Biomass-Degrading Enzymes, Conidiation, and Pigment Biosynthesis in Penicillium oxalicum
Abstract
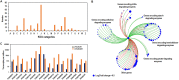
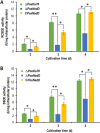
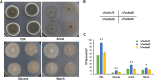
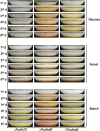
Soil fungi produce a wide range of chemical compounds and enzymes with potential for applications in medicine and biotechnology. Cellular processes in soil fungi are highly dependent on the regulation under environmentally induced stress, but most of the underlying mechanisms remain unclear. Previous work identified a key GATA-type transcription factor, Penicillium oxalicum NsdD (PoxNsdD; also called POX08415), that regulates the expression of cellulase and xylanase genes in P. oxalicum PoxNsdD shares 57 to 64% identity with the key activator NsdD, involved in asexual development in Aspergillus In the present study, the regulatory roles of PoxNsdD in P. oxalicum were further explored. Comparative transcriptomic profiling revealed that PoxNsdD regulates major genes involved in starch, cellulose, and hemicellulose degradation, as well as conidiation and pigment biosynthesis. Subsequent experiments confirmed that a ΔPoxNsdD strain lost 43.9 to 78.8% of starch-digesting enzyme activity when grown on soluble corn starch, and it produced 54.9 to 146.0% more conidia than the ΔPoxKu70 parental strain. During cultivation, ΔPoxNsdD cultures changed color, from pale orange to brick red, while the ΔPoxKu70 cultures remained bluish white. Real-time quantitative reverse transcription-PCR showed that PoxNsdD dynamically regulated the expression of a glucoamylase gene (POX01356/Amy15A), an α-amylase gene (POX09352/Amy13A), and a regulatory gene (POX03890/amyR), as well as a polyketide synthase gene (POX01430/alb1/wA) for yellow pigment biosynthesis and a conidiation-regulated gene (POX06534/brlA). Moreover, in vitro binding experiments showed that PoxNsdD bound the promoter regions of the above-described genes. This work provides novel insights into the regulatory mechanisms of fungal cellular processes and may assist in genetic engineering of Poxalicum for potential industrial and medical applications.IMPORTANCE Most filamentous fungi produce a vast number of extracellular enzymes that are used commercially for biorefineries of plant biomass to produce biofuels and value-added chemicals, which might promote the transition to a more environmentally friendly economy. The expression of these extracellular enzyme genes is tightly controlled at the transcriptional level, which limits their yields. Hitherto our understanding of the regulation of expression of plant biomass-degrading enzyme genes in filamentous fungi has been rather limited. In the present study, regulatory roles of a key regulator, PoxNsdD, were further explored in the soil fungus Penicillium oxalicum, contributing to the understanding of gene regulation in filamentous fungi and revealing the biotechnological potential of Poxalicum via genetic engineering.
Keywords: Penicillium oxalicum; PoxNsdD; conidiation; pigment biosynthesis; starch-degrading enzyme; transcription factor.
Copyright © 2018 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

