Regulation of PI4P levels by PI4KIIIα during G-protein-coupled PLC signaling in Drosophila photoreceptors
- PMID: 29980590
- PMCID: PMC6104824
- DOI: 10.1242/jcs.217257
Regulation of PI4P levels by PI4KIIIα during G-protein-coupled PLC signaling in Drosophila photoreceptors
Abstract
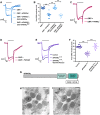
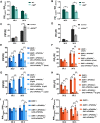
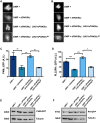
The activation of phospholipase C (PLC) is a conserved mechanism of receptor-activated cell signaling at the plasma membrane. PLC hydrolyzes the minor membrane lipid phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], and continued signaling requires the resynthesis and availability of PI(4,5)P2 at the plasma membrane. PI(4,5)P2 is synthesized by the phosphorylation of phosphatidylinositol 4-phosphate (PI4P). Thus, a continuous supply of PI4P is essential to support ongoing PLC signaling. While the enzyme PI4KA has been identified as performing this function in cultured mammalian cells, its function in the context of an in vivo physiological model has not been established. In this study, we show that, in Drosophila photoreceptors, PI4KIIIα activity is required to support signaling during G-protein-coupled PLC activation. Depletion of PI4KIIIα results in impaired electrical responses to light, and reduced plasma membrane levels of PI4P and PI(4,5)P2 Depletion of the conserved proteins Efr3 and TTC7 [also known as StmA and L(2)k14710, respectively, in flies], which assemble PI4KIIIα at the plasma membrane, also results in an impaired light response and reduced plasma membrane PI4P and PI(4,5)P2 levels. Thus, PI4KIIIα activity at the plasma membrane generates PI4P and supports PI(4,5)P2 levels during receptor activated PLC signaling.
Keywords: Drosophila; PI4KIIIα; PI4P; PLC.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
-
- Balla A., Tuymetova G., Tsiomenko A., Várnai P. and Balla T. (2005). A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell 16, 1282-1295. 10.1091/mbc.e04-07-0578 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

