A mosaic monoploid reference sequence for the highly complex genome of sugarcane
- PMID: 29980662
- PMCID: PMC6035169
- DOI: 10.1038/s41467-018-05051-5
A mosaic monoploid reference sequence for the highly complex genome of sugarcane
Abstract
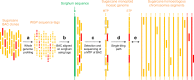
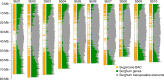
Sugarcane (Saccharum spp.) is a major crop for sugar and bioenergy production. Its highly polyploid, aneuploid, heterozygous, and interspecific genome poses major challenges for producing a reference sequence. We exploited colinearity with sorghum to produce a BAC-based monoploid genome sequence of sugarcane. A minimum tiling path of 4660 sugarcane BAC that best covers the gene-rich part of the sorghum genome was selected based on whole-genome profiling, sequenced, and assembled in a 382-Mb single tiling path of a high-quality sequence. A total of 25,316 protein-coding gene models are predicted, 17% of which display no colinearity with their sorghum orthologs. We show that the two species, S. officinarum and S. spontaneum, involved in modern cultivars differ by their transposable elements and by a few large chromosomal rearrangements, explaining their distinct genome size and distinct basic chromosome numbers while also suggesting that polyploidization arose in both lineages after their divergence.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- European Commission of Agriculture and rural development. Sugar. http://ec.europa.eu/agriculture/sugar/index_en.htm.
-
- Gouy, M., Nibouche, S., Hoarau, J. H., & Costet, L. in Translational Genomics for Crop Breeding: Abiotic Stress, Yield and Quality, Vol. 2, (eds. Varshney, R. & Tuberosa, R.) Ch. 13, 211–237 (John Wiley & Sons, Ames, Chichester and Oxford, 2013).
-
- D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome. 1998;41:221–225. doi: 10.1139/g98-023. - DOI
-
- Screenivasan, T. V., Ahloowalia, B. S. & Heinz, D. J. Sugarcane improvement through breeding. Cytogenetics5, 211–253 (1987).
-
- Brandes E. Origin, dispersal and use in breeding of the Melanesian garden sugarcane and their derivatives, Saccharum officinarum L. Proc. Int. Soc. Sugarcane Technol. 1956;9:709–750.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

