Exploiting limited valence patchy particles to understand autocatalytic kinetics
- PMID: 29980675
- PMCID: PMC6035234
- DOI: 10.1038/s41467-018-04977-0
Exploiting limited valence patchy particles to understand autocatalytic kinetics
Abstract
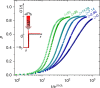
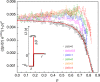
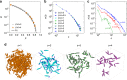
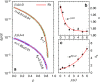
Autocatalysis, i.e., the speeding up of a reaction through the very same molecule which is produced, is common in chemistry, biophysics, and material science. Rate-equation-based approaches are often used to model the time dependence of products, but the key physical mechanisms behind the reaction cannot be properly recognized. Here, we develop a patchy particle model inspired by a bicomponent reactive mixture and endowed with adjustable autocatalytic ability. Such a coarse-grained model captures all general features of an autocatalytic aggregation process that takes place under controlled and realistic conditions, including crowded environments. Simulation reveals that a full understanding of the kinetics involves an unexpected effect that eludes the chemistry of the reaction, and which is crucially related to the presence of an activation barrier. The resulting analytical description can be exported to real systems, as confirmed by experimental data on epoxy-amine polymerizations, solving a long-standing issue in their mechanistic description.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Universal motifs and the diversity of autocatalytic systems.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25230-25236. doi: 10.1073/pnas.2013527117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989134 Free PMC article.
-
Stoechiometric and dynamical autocatalysis for diluted chemical reaction networks.J Math Biol. 2022 Sep 7;85(3):26. doi: 10.1007/s00285-022-01798-0. J Math Biol. 2022. PMID: 36071258
-
Kinetics of autocatalysis in small systems.J Chem Phys. 2008 Jan 7;128(1):015101. doi: 10.1063/1.2815800. J Chem Phys. 2008. PMID: 18190223
-
Unified representation of Life's basic properties by a 3-species Stochastic Cubic Autocatalytic Reaction-Diffusion system of equations.Phys Life Rev. 2022 Jul;41:64-83. doi: 10.1016/j.plrev.2022.03.003. Epub 2022 May 13. Phys Life Rev. 2022. PMID: 35594602 Review.
-
Colloidal aspects of digestion of Pickering emulsions: Experiments and theoretical models of lipid digestion kinetics.Adv Colloid Interface Sci. 2019 Jan;263:195-211. doi: 10.1016/j.cis.2018.10.002. Epub 2018 Oct 22. Adv Colloid Interface Sci. 2019. PMID: 30580767 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

