Glycan recognition in globally dominant human rotaviruses
- PMID: 29980685
- PMCID: PMC6035239
- DOI: 10.1038/s41467-018-05098-4
Glycan recognition in globally dominant human rotaviruses
Abstract
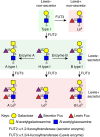
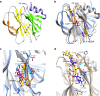
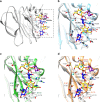
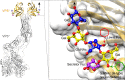
Rotaviruses (RVs) cause life-threatening diarrhea in infants and children worldwide. Recent biochemical and epidemiological studies underscore the importance of histo-blood group antigens (HBGA) as both cell attachment and susceptibility factors for the globally dominant P[4], P[6], and P[8] genotypes of human RVs. How these genotypes interact with HBGA is not known. Here, our crystal structures of P[4] and a neonate-specific P[6] VP8*s alone and in complex with H-type I HBGA reveal a unique glycan binding site that is conserved in the globally dominant genotypes and allows for the binding of ABH HBGAs, consistent with their prevalence. Remarkably, the VP8* of P[6] RVs isolated from neonates displays subtle structural changes in this binding site that may restrict its ability to bind branched glycans. This provides a structural basis for the age-restricted tropism of some P[6] RVs as developmentally regulated unbranched glycans are more abundant in the neonatal gut.
Conflict of interest statement
The authors declare no competing interests.
Figures


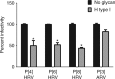

References
-
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization-Coordinated Global Rotavirus Surveillance Network Global, regional, and national estimates of rotavirus mortality in children<5 years of age, 2000-2013. Clin. Infect. Dis. 2016;62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. - DOI - PMC - PubMed
-
- Estes, M. K. & Greenberg, H. B. in Fields Virology (eds Knipe, D. M. et al.) 1347–1401 (Lippincott Williams & Wilkins, Philadelphia, PA, 2013).
Publication types
MeSH terms
Substances
Grants and funding
- AI36040/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)/International
- R01 AI105101/AI/NIAID NIH HHS/United States
- P30 GM124169/GM/NIGMS NIH HHS/United States
- R56 AI080656/AI/NIAID NIH HHS/United States
- AI 080656 and AI 105101/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)/International
- GM098791 and P41GM103694/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R01 AI036040/AI/NIAID NIH HHS/United States
- R01 AI080656/AI/NIAID NIH HHS/United States
- R37 AI036040/AI/NIAID NIH HHS/United States
- P30 DK056338/DK/NIDDK NIH HHS/United States
- P41 GM103694/GM/NIGMS NIH HHS/United States
- R24 GM098791/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

