Gestational exposure to chlordecone promotes transgenerational changes in the murine reproductive system of males
- PMID: 29980752
- PMCID: PMC6035262
- DOI: 10.1038/s41598-018-28670-w
Gestational exposure to chlordecone promotes transgenerational changes in the murine reproductive system of males
Abstract
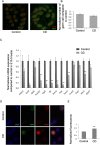
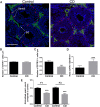
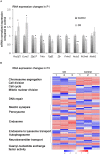
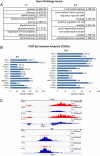
Environmental factors can affect epigenetic events during germline reprogramming and impose distinctive transgenerational consequences onto the offspring. In this study, we examined the transgenerational effects of chlordecone (CD), an organochlorine insecticide with well-known estrogenic properties. We exposed pregnant mice to CD from embryonic day 6.5 to 15.5 and observed a reduction in spermatogonia (SG) numbers in F3, meiotic defects in spermatocytes and decrease in spermatozoa number in the first and third generation of male progeny. The RNA qRT-PCR expression analysis in F1 and transcriptomics analysis in F3 males using the whole testes revealed changes in the expression of genes associated with chromosome segregation, cell division and DNA repair. The expression of the master regulator of pluripotency, Pou5f1, decreased in foetal and increased in adult F1, but not in F3 adult testes. Analysis of histone H3K4me3 distribution revealed widespread changes in its occupancy in the genome of F1 and F3 generations. We established that 7.1% of altered epigenetic marks were conserved between F1 and F3 generations. The overlapping changes common to F1 and F3 include genes implicated in cell adhesion and transcription factor activities functions. Differential peaks observed in F1 males are significantly enriched in predicted ESR1 binding sites, some of which we confirmed to be functional. Our data demonstrate that CD-mediated impairment of reproductive functions could be transmitted to subsequent generations.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16061–16066. doi: 10.1073/pnas.1315204110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

