Genome Evolution of Bartonellaceae Symbionts of Ants at the Opposite Ends of the Trophic Scale
- PMID: 29982531
- PMCID: PMC6044324
- DOI: 10.1093/gbe/evy126
Genome Evolution of Bartonellaceae Symbionts of Ants at the Opposite Ends of the Trophic Scale
Abstract
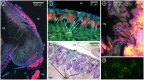
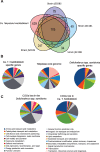
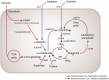
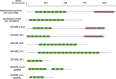
Many insects rely on bacterial symbionts to supply essential amino acids and vitamins that are deficient in their diets, but metabolic comparisons of closely related gut bacteria in insects with different dietary preferences have not been performed. Here, we demonstrate that herbivorous ants of the genus Dolichoderus from the Peruvian Amazon host bacteria of the family Bartonellaceae, known for establishing chronic or pathogenic infections in mammals. We detected these bacteria in all studied Dolichoderus species, and found that they reside in the midgut wall, that is, the same location as many previously described nutritional endosymbionts of insects. The genomic analysis of four divergent strains infecting different Dolichoderus species revealed genes encoding pathways for nitrogen recycling and biosynthesis of several vitamins and all essential amino acids. In contrast, several biosynthetic pathways have been lost, whereas genes for the import and conversion of histidine and arginine to glutamine have been retained in the genome of a closely related gut bacterium of the carnivorous ant Harpegnathos saltator. The broad biosynthetic repertoire in Bartonellaceae of herbivorous ants resembled that of gut bacteria of honeybees that likewise feed on carbohydrate-rich diets. Taken together, the broad distribution of Bartonellaceae across Dolichoderus ants, their small genome sizes, the specific location within hosts, and the broad biosynthetic capability suggest that these bacteria are nutritional symbionts in herbivorous ants. The results highlight the important role of the host nutritional biology for the genomic evolution of the gut microbiota-and conversely, the importance of the microbiota for the nutrition of hosts.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Anderson KE, et al. . 2012. Highly similar microbial communities are shared among related and trophically similar ant species. Mol Ecol. 21(9):2282–2296. - PubMed
-
- Baker GC, Smith JJ, Cowan DA.. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55(3):541–555. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

