Coupled catabolism and anabolism in autocatalytic RNA sets
- PMID: 29982824
- PMCID: PMC6182175
- DOI: 10.1093/nar/gky598
Coupled catabolism and anabolism in autocatalytic RNA sets
Abstract
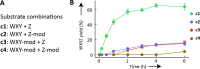
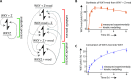
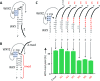
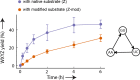
The ability to process molecules available in the environment into useable building blocks characterizes catabolism in contemporary cells and was probably critical for the initiation of life. Here we show that a catabolic process in collectively autocatalytic sets of RNAs allows diversified substrates to be assimilated. We modify fragments of the Azoarcus group I intron and find that the system is able to restore the original native fragments by a multi-step reaction pathway. This allows in turn the formation of catalysts by an anabolic process, eventually leading to the accumulation of ribozymes. These results demonstrate that rudimentary self-reproducing RNA systems based on recombination possess an inherent capacity to assimilate an expanded repertoire of chemical resources and suggest that coupled catabolism and anabolism could have arisen at a very early stage in primordial living systems.
Figures

References
-
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971; 58:465–523. - PubMed
-
- Eigen M., Schuster P.. The hypercycle. A principle of natural self-organization. Part A: emergence of the hypercycle. Naturwissenschaften. 1977; 64:541–565. - PubMed
-
- Kauffman S.A. Autocatalytic sets of proteins. J. Theor. Biol. 1986; 119:1–24. - PubMed
-
- Higgs P.G., Lehman N.. The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 2015; 16:7–17. - PubMed
-
- Hordijk W., Steel M.. Chasing the tail: the emergence of autocatalytic networks. Biosystems. 2017; 152:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

