Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics
- PMID: 29983054
- PMCID: PMC6070396
- DOI: 10.1021/jacs.8b03896
Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics
Abstract
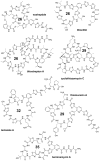
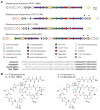
Thiopeptides are members of the ribosomally synthesized and post-translationally modified peptide family of natural products. Most characterized thiopeptides display nanomolar potency toward Gram-positive bacteria by blocking protein translation with several being produced at the industrial scale for veterinary and livestock applications. Employing our custom bioinformatics program, RODEO, we expand the thiopeptide family of natural products by a factor of four. This effort revealed many new thiopeptide biosynthetic gene clusters with products predicted to be distinct from characterized thiopeptides and identified gene clusters for previously characterized molecules of unknown biosynthetic origin. To further validate our data set of predicted thiopeptide biosynthetic gene clusters, we isolated and characterized a structurally unique thiopeptide featuring a central piperidine and rare thioamide moiety. Termed saalfelduracin, this thiopeptide displayed potent antibiotic activity toward several drug-resistant Gram-positive pathogens. A combination of whole-genome sequencing, comparative genomics, and heterologous expression experiments confirmed that the thioamide moiety of saalfelduracin is installed post-translationally by the joint action of two proteins, TfuA and YcaO. These results reconcile the previously unknown origin of the thioamide in two long-known thiopeptides, thiopeptin and Sch 18640. Armed with these new insights into thiopeptide chemical-genomic space, we provide a roadmap for the discovery of additional members of this natural product family.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30:108. - PMC - PubMed
-
- Bagley MC, Dale JW, Merritt EA, Xiong X. Chem Rev. 2005;105:685. - PubMed
-
- Hashimoto M, Murakami T, Funahashi K, Tokunaga T, Nihei K-i, Okuno T, Kimura T, Naoki H, Himeno H. Bioorganic Med Chem. 2006;14:8259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

