De Novo Mutation in Genes Regulating Neural Stem Cell Fate in Human Congenital Hydrocephalus
- PMID: 29983323
- PMCID: PMC7839075
- DOI: 10.1016/j.neuron.2018.06.019
De Novo Mutation in Genes Regulating Neural Stem Cell Fate in Human Congenital Hydrocephalus
Abstract
Congenital hydrocephalus (CH), featuring markedly enlarged brain ventricles, is thought to arise from failed cerebrospinal fluid (CSF) homeostasis and is treated with lifelong surgical CSF shunting with substantial morbidity. CH pathogenesis is poorly understood. Exome sequencing of 125 CH trios and 52 additional probands identified three genes with significant burden of rare damaging de novo or transmitted mutations: TRIM71 (p = 2.15 × 10-7), SMARCC1 (p = 8.15 × 10-10), and PTCH1 (p = 1.06 × 10-6). Additionally, two de novo duplications were identified at the SHH locus, encoding the PTCH1 ligand (p = 1.2 × 10-4). Together, these probands account for ∼10% of studied cases. Strikingly, all four genes are required for neural tube development and regulate ventricular zone neural stem cell fate. These results implicate impaired neurogenesis (rather than active CSF accumulation) in the pathogenesis of a subset of CH patients, with potential diagnostic, prognostic, and therapeutic ramifications.
Keywords: PTCH1; SHH; SMARCC1; TRIM71; aqueductal stenosis; congenital hydrocephalus; de novo variants; gene discovery; neural stem cell; whole-exome sequencing.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
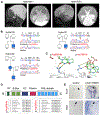
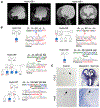

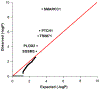
Comment in
-
Genetics Sheds New Light on Congenital Hydrocephalus Biology.Neuron. 2018 Jul 25;99(2):246-247. doi: 10.1016/j.neuron.2018.07.008. Neuron. 2018. PMID: 30048611
References
-
- Adle-Biassette H, Saugier-Veber P, Fallet-Bianco C, Delezoide A-L, Razavi F, Drouot N, Bazin A, Beaufrère A-M, Bessières B, Blesson S, et al. (2013). Neuropathological review of 138 cases genetically tested for X-linked hydrocephalus: evidence for closely related clinical entities of unknown molecular bases. Acta Neuropathol. 126, 427–442. - PubMed
-
- Aeschimann F, Kumari P, Bartake H, Gaidatzis D, Xu L, Ciosk R, and Großhans H (2017). LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell 65, 476–489.e4. - PubMed
-
- Al-Dosari MS, Al-Owain M, Tulbah M, Kurdi W, Adly N, Al-Hemidan A, Masoodi TA, Albash B, and Alkuraya FS (2013). Mutation in MPDZ causes severe congenital hydrocephalus. J. Med. Genet 50, 54–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

