Cancer-predicting transcriptomic and epigenetic signatures revealed for ulcerative colitis in patient-derived epithelial organoids
- PMID: 29983891
- PMCID: PMC6033374
- DOI: 10.18632/oncotarget.25617
Cancer-predicting transcriptomic and epigenetic signatures revealed for ulcerative colitis in patient-derived epithelial organoids
Abstract
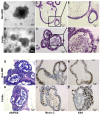
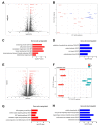
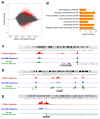
Ulcerative colitis (UC) is a prevalent form of inflammatory bowel disease (IBD) whose pathogenic mechanisms remain unclear. Elucidating these mechanisms is important to reduce UC symptoms and to prevent UC progression into colitis-associated colon cancer (CAC). Our goal was to develop and validate faithful, human-derived, UC models and analyze them at histologic, transcriptomic and epigenetic levels to allow mechanistic studies of UC and CAC pathogenesis. We generated patient-derived primary-organoid cultures from UC and non-IBD colonic epithelium. We phenotyped them histologically and used next-generation-sequencing approaches to profile whole transcriptomes and epigenomes of organoids and primary tissues. Tissue organization and expression of mucin 2 (MUC2) and lysozyme (LYZ) demonstrated histologic faithfulness of organoids to healthy and diseased colonic epithelium. Transcriptomic analyses showed increased expression of inflammatory pathways in UC patient-derived organoids and tissues. Profiling for active enhancers using the H3K27ac histone modification revealed UC-derived organoid enrichment for pathways indicative of gastrointestinal cancer, including S100 calcium-binding protein P (S100P), and revealed novel markers for GI cancer, including both LYZ and neuropeptide S receptor 1 (NPSR1). Immunolocalization showed increased levels of LYZ, S100P, and NPSR1 proteins in UC and CAC. In conclusion, primary colonic organoid cultures from UC and non-IBD patients can be established that faithfully represent diseased or normal colonic states. These models reveal precancerous molecular pathways that are already activated in UC. The findings demonstrate the suitability of primary organoids for dissecting UC and CAC pathogenic mechanisms and suggest new targets for therapeutic intervention.
Keywords: H3K27Ac enhancer chromatin mark; colitis associated cancer; inflammation; organoids; ulcerative colitis.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures

References
-
- Dotti I, Mora-Buch R, Ferrer-Picon E, Planell N, Jung P, Masamunt MC, Leal RF, Martin de Carpi J, Llach J, Ordas I, Batlle E, Panes J, Salas A. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–79. doi: 10.1136/gutjnl-2016-312609. - DOI - PMC - PubMed
-
- Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. - DOI - PMC - PubMed
Grants and funding
- TL1 TR002549/TR/NCATS NIH HHS/United States
- R01 AR068308/AR/NIAMS NIH HHS/United States
- P50 CA150964/CA/NCI NIH HHS/United States
- TL1 TR000441/TR/NCATS NIH HHS/United States
- P01 HL029582/HL/NHLBI NIH HHS/United States
- UL1 TR000439/TR/NCATS NIH HHS/United States
- U01 CA214300/CA/NCI NIH HHS/United States
- P01 CA062220/CA/NCI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R01 CA193359/CA/NCI NIH HHS/United States
- R01 CA157663/CA/NCI NIH HHS/United States
- P01 HL076491/HL/NHLBI NIH HHS/United States
- R01 CA142808/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

