Gut microbiota associations with common diseases and prescription medications in a population-based cohort
- PMID: 29985401
- PMCID: PMC6037668
- DOI: 10.1038/s41467-018-05184-7
Gut microbiota associations with common diseases and prescription medications in a population-based cohort
Abstract
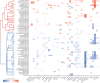
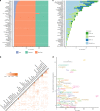
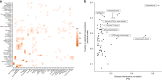
The human gut microbiome has been associated with many health factors but variability between studies limits exploration of effects between them. Gut microbiota profiles are available for >2700 members of the deeply phenotyped TwinsUK cohort, providing a uniform platform for such comparisons. Here, we present gut microbiota association analyses for 38 common diseases and 51 medications within the cohort. We describe several novel associations, highlight associations common across multiple diseases, and determine which diseases and medications have the greatest association with the gut microbiota. These results provide a reference for future studies of the gut microbiome and its role in human health.
Conflict of interest statement
T.D.S. is co-founder of MapMySelf and MapMyGut Ltd. The remaining authors declare no competing interests.
Figures

Comment in
-
Response to: Population-Based Gut Microbiome Associations With Hypertension.Circ Res. 2018 Nov 9;123(11):1188-1189. doi: 10.1161/CIRCRESAHA.118.313913. Circ Res. 2018. PMID: 30571463 No abstract available.
-
Population-Based Gut Microbiome Associations With Hypertension.Circ Res. 2018 Nov 9;123(11):1185-1187. doi: 10.1161/CIRCRESAHA.118.313792. Circ Res. 2018. PMID: 30571464 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

