Data-driven analysis of motor activity implicates 5-HT2A neurons in backward locomotion of larval Drosophila
- PMID: 29985473
- PMCID: PMC6037780
- DOI: 10.1038/s41598-018-28680-8
Data-driven analysis of motor activity implicates 5-HT2A neurons in backward locomotion of larval Drosophila
Abstract
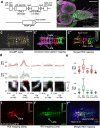
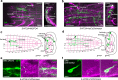
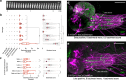
Rhythmic animal behaviors are regulated in part by neural circuits called the central pattern generators (CPGs). Classifying neural population activities correlated with body movements and identifying the associated component neurons are critical steps in understanding CPGs. Previous methods that classify neural dynamics obtained by dimension reduction algorithms often require manual optimization which could be laborious and preparation-specific. Here, we present a simpler and more flexible method that is based on the pre-trained convolutional neural network model VGG-16 and unsupervised learning, and successfully classifies the fictive motor patterns in Drosophila larvae under various imaging conditions. We also used voxel-wise correlation mapping to identify neurons associated with motor patterns. By applying these methods to neurons targeted by 5-HT2A-GAL4, which we generated by the CRISPR/Cas9-system, we identified two classes of interneurons, termed Seta and Leta, which are specifically active during backward but not forward fictive locomotion. Optogenetic activation of Seta and Leta neurons increased backward locomotion. Conversely, thermogenetic inhibition of 5-HT2A-GAL4 neurons or application of a 5-HT2 antagonist decreased backward locomotion induced by noxious light stimuli. This study establishes an accelerated pipeline for activity profiling and cell identification in larval Drosophila and implicates the serotonergic system in the modulation of backward locomotion.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Gap Junction-Mediated Signaling from Motor Neurons Regulates Motor Generation in the Central Circuits of Larval Drosophila.J Neurosci. 2017 Feb 22;37(8):2045-2060. doi: 10.1523/JNEUROSCI.1453-16.2017. Epub 2017 Jan 23. J Neurosci. 2017. PMID: 28115483 Free PMC article.
-
Identification of Inhibitory Premotor Interneurons Activated at a Late Phase in a Motor Cycle during Drosophila Larval Locomotion.PLoS One. 2015 Sep 3;10(9):e0136660. doi: 10.1371/journal.pone.0136660. eCollection 2015. PLoS One. 2015. PMID: 26335437 Free PMC article.
-
Functional Genetic Screen to Identify Interneurons Governing Behaviorally Distinct Aspects of Drosophila Larval Motor Programs.G3 (Bethesda). 2016 Jul 7;6(7):2023-31. doi: 10.1534/g3.116.028472. G3 (Bethesda). 2016. PMID: 27172197 Free PMC article.
-
Neural Circuits Underlying Fly Larval Locomotion.Curr Pharm Des. 2017;23(12):1722-1733. doi: 10.2174/1381612822666161208120835. Curr Pharm Des. 2017. PMID: 27928962 Free PMC article. Review.
-
Optogenetics in Drosophila.Adv Exp Med Biol. 2021;1293:309-320. doi: 10.1007/978-981-15-8763-4_19. Adv Exp Med Biol. 2021. PMID: 33398822 Review.
Cited by
-
Regulation of coordinated muscular relaxation in Drosophila larvae by a pattern-regulating intersegmental circuit.Nat Commun. 2021 May 19;12(1):2943. doi: 10.1038/s41467-021-23273-y. Nat Commun. 2021. PMID: 34011945 Free PMC article.
-
Molecular Tools for Targeted Control of Nerve Cell Electrical Activity. Part II.Acta Naturae. 2021 Oct-Dec;13(4):17-32. doi: 10.32607/actanaturae.11415. Acta Naturae. 2021. PMID: 35127143 Free PMC article.
-
An Iterative Method for Predicting Essential Proteins Based on Multifeature Fusion and Linear Neighborhood Similarity.Front Aging Neurosci. 2022 Jan 24;13:799500. doi: 10.3389/fnagi.2021.799500. eCollection 2021. Front Aging Neurosci. 2022. PMID: 35140599 Free PMC article.
-
A pair of ascending neurons in the subesophageal zone mediates aversive sensory inputs-evoked backward locomotion in Drosophila larvae.PLoS Genet. 2020 Nov 2;16(11):e1009120. doi: 10.1371/journal.pgen.1009120. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137117 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

