Predicting Cotranscriptional Folding Kinetics For Riboswitch
- PMID: 29985608
- PMCID: PMC6345277
- DOI: 10.1021/acs.jpcb.8b04249
Predicting Cotranscriptional Folding Kinetics For Riboswitch
Abstract
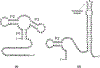
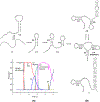
On the basis of a helix-based transition rate model, we developed a new method for sampling cotranscriptional RNA conformational ensemble and the prediction of cotranscriptional folding kinetics. Applications to E. coli. SRP RNA and pbuE riboswitch indicate that the model may provide reliable predictions for the cotranscriptional folding pathways and population kinetics. For E. coli. SRP RNA, the predicted population kinetics and the folding pathway are consistent with the SHAPE profiles in the recent cotranscriptional SHAPE-seq experiments. For the pbuE riboswitch, the model predicts the transcriptional termination efficiency as a function of the force. The theoretical results show (a) a force-induced transition from the aptamer (antiterminator) to the terminator structure and (b) the different folding pathways for the riboswitch with and without the ligand (adenine). More specifically, without adenine, the aptamer structure emerges as a short-lived kinetic transient state instead of a thermodynamically stable intermediate state. Furthermore, from the predicted extension-time curves, the model identifies a series of conformational switches in the pulling process, where the predicted relative residence times for the different structures are in accordance with the experimental data. The model may provide a new tool for quantitative predictions of cotranscriptional folding kinetics, and results can offer useful insights into cotranscriptional folding-related RNA functions such as regulation of gene expression with riboswitches.
Figures
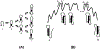
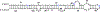

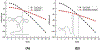
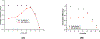
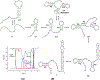

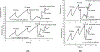
Similar articles
-
The regulation mechanism of yitJ and metF riboswitches.J Chem Phys. 2015 Jul 28;143(4):045103. doi: 10.1063/1.4927390. J Chem Phys. 2015. PMID: 26233166
-
Single-molecule FRET studies on the cotranscriptional folding of a thiamine pyrophosphate riboswitch.Proc Natl Acad Sci U S A. 2018 Jan 9;115(2):331-336. doi: 10.1073/pnas.1712983115. Epub 2017 Dec 26. Proc Natl Acad Sci U S A. 2018. PMID: 29279370 Free PMC article.
-
Cotranscriptional folding of a riboswitch at nucleotide resolution.Nat Struct Mol Biol. 2016 Dec;23(12):1124-1131. doi: 10.1038/nsmb.3316. Epub 2016 Oct 31. Nat Struct Mol Biol. 2016. PMID: 27798597 Free PMC article.
-
Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design.Wiley Interdiscip Rev RNA. 2015 Nov-Dec;6(6):631-50. doi: 10.1002/wrna.1300. Epub 2015 Sep 11. Wiley Interdiscip Rev RNA. 2015. PMID: 26361734 Free PMC article. Review.
-
Computational Methods for Modeling Aptamers and Designing Riboswitches.Int J Mol Sci. 2017 Nov 17;18(11):2442. doi: 10.3390/ijms18112442. Int J Mol Sci. 2017. PMID: 29149090 Free PMC article. Review.
Cited by
-
cRNAsp12 Web Server for the Prediction of Circular RNA Secondary Structures and Stabilities.Int J Mol Sci. 2023 Feb 14;24(4):3822. doi: 10.3390/ijms24043822. Int J Mol Sci. 2023. PMID: 36835231 Free PMC article.
-
Observation of coordinated RNA folding events by systematic cotranscriptional RNA structure probing.Nat Commun. 2023 Nov 29;14(1):7839. doi: 10.1038/s41467-023-43395-9. Nat Commun. 2023. PMID: 38030633 Free PMC article.
-
Requirements for efficient ligand-gated co-transcriptional switching in designed variants of the B. subtilis pbuE adenine-responsive riboswitch in E. coli.PLoS One. 2020 Dec 1;15(12):e0243155. doi: 10.1371/journal.pone.0243155. eCollection 2020. PLoS One. 2020. PMID: 33259551 Free PMC article.
-
Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity.Chem Rev. 2021 Jul 14;121(13):7398-7467. doi: 10.1021/acs.chemrev.1c00009. Epub 2021 May 26. Chem Rev. 2021. PMID: 34038115 Free PMC article. Review.
-
Predicting the Structure of a Viroid : Structure, Structure Distribution, Consensus Structure, and Structure Drawing.Methods Mol Biol. 2022;2316:331-371. doi: 10.1007/978-1-0716-1464-8_26. Methods Mol Biol. 2022. PMID: 34845705
References
-
- Boyle J; Robillard GT; Kim SH Sequential Folding of Transfer RNA: A Nuclear Magnetic Resonance Study of Successively Longer tRNA Fragments With a Common 5 End. J. Mol. Biol 1980, 139, 601–625. - PubMed
-
- Sosnick TR; Pan T RNA Folding During Transcription. Annu. Rev. Biophys. Biomol. Struct 2006,35,161–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

