High-depth transcriptomic profiling reveals the temporal gene signature of human mesenchymal stem cells during chondrogenesis
- PMID: 29985644
- PMCID: PMC6355072
- DOI: 10.1096/fj.201800534R
High-depth transcriptomic profiling reveals the temporal gene signature of human mesenchymal stem cells during chondrogenesis
Abstract
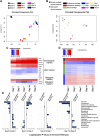
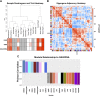
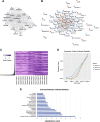
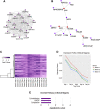
Mesenchymal stem/stromal cells (MSCs) provide an attractive cell source for cartilage repair and cell therapy; however, the underlying molecular pathways that drive chondrogenesis of these populations of adult stem cells remain poorly understood. We generated a rich data set of high-throughput RNA sequencing of human MSCs throughout chondrogenesis at 6 different time points. Our data consisted of 18 libraries with 3 individual donors as biologic replicates, with each library possessing a sequencing depth of 100 million reads. Computational analyses with differential gene expression, gene ontology, and weighted gene correlation network analysis identified dynamic changes in multiple biologic pathways and, most importantly, a chondrogenic gene subset, whose functional characterization promises to further harness the potential of MSCs for cartilage tissue engineering. Furthermore, we created a graphic user interface encyclopedia built with the goal of producing an open resource of transcriptomic regulation for additional data mining and pathway analysis of the process of MSC chondrogenesis.-Huynh, N. P. T., Zhang, B., Guilak, F. High-depth transcriptomic profiling reveals the temporal gene signature of human mesenchymal stem cells during chondrogenesis.
Keywords: RNA-Seq; chondrocyte; lncRNA; miRNA; pericyte.
Conflict of interest statement
The authors thank the Duke University Genomics Core Facility, the Washington University in St. Louis Pathology Core Labs, and the Washington University Center of Regenerative Medicine for their resources and support. The authors also wish to thank S. Oswald (Washington University in St. Louis) for assistance with technical writing. This work was supported by the Arthritis Foundation, U.S. National Institutes of Health (NIH) Grants AR50245, AR48852, AG15768, AR48182, and AR067467, the Nancy Taylor Foundation for Chronic Diseases, the Collaborative Research Center of the AO Foundation, Davos, Switzerland, and the Washington University Musculoskeletal Research Center (NIH P30 AR057235). B.Z. was supported by NIH grant DA027995 and the Goldman Sachs Philanthropy Fund. The authors declare no conflicts of interest.
Figures


References
-
- Ahmed T. A., Hincke M. T. (2014) Mesenchymal stem cell–based tissue engineering strategies for repair of articular cartilage. Histol. Histopathol. 29, 669–689 - PubMed
-
- Crisan M., Yap S., Casteilla L., Chen C. W., Corselli M., Park T. S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P. N., Traas J., Schugar R., Deasy B. M., Badylak S., Buhring H. J., Giacobino J. P., Lazzari L., Huard J., Péault B. (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 10.1016/j.stem.2008.07.003 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

