Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey
- PMID: 29987016
- PMCID: PMC6065006
- DOI: 10.1073/pnas.1803573115
Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey
Abstract
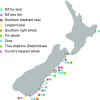
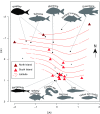
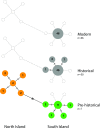
New Zealand's geographic isolation, lack of native terrestrial mammals, and Gondwanan origins make it an ideal location to study evolutionary processes. However, since the archipelago was first settled by humans 750 y ago, its unique biodiversity has been under pressure, and today an estimated 49% of the terrestrial avifauna is extinct. Current efforts to conserve the remaining fauna rely on a better understanding of the composition of past ecosystems, as well as the causes and timing of past extinctions. The exact temporal and spatial dynamics of New Zealand's extinct fauna, however, can be difficult to interpret, as only a small proportion of animals are preserved as morphologically identifiable fossils. Here, we conduct a large-scale genetic survey of subfossil bone assemblages to elucidate the impact of humans on the environment in New Zealand. By genetically identifying more than 5,000 nondiagnostic bone fragments from archaeological and paleontological sites, we reconstruct a rich faunal record of 110 species of birds, fish, reptiles, amphibians, and marine mammals. We report evidence of five whale species rarely reported from New Zealand archaeological middens and characterize extinct lineages of leiopelmatid frog (Leiopelma sp.) and kākāpō (Strigops habroptilus) haplotypes lost from the gene pool. Taken together, this molecular audit of New Zealand's subfossil record not only contributes to our understanding of past biodiversity and precontact Māori subsistence practices but also provides a more nuanced snapshot of anthropogenic impacts on native fauna after first human arrival.
Keywords: ancient DNA; bulk bone metabarcoding; human impacts; paleoecology; subsistence practices.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Tennyson AJD. The origin and history of New Zealand’s terrestrial vertebrates. N Z J Ecol. 2010;34:6–27.
-
- Anderson A. Changing perspectives upon Māori colonisation voyaging. J R Soc N Z. 2017;47:222–231.
-
- Duncan RP, Blackburn TM. Extinction and endemism in New Zealand land birds. Glob Ecol Biogeogr. 2004;13:509–517.
-
- Worthy TH, Holdaway RN. The Lost World of the Moa: Prehistoric Life of New Zealand. Indiana Univ Press; Bloomington, IN: 2002.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Dryad/10.5061/dryad.ss0k19q
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

