A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer disease
- PMID: 29988083
- PMCID: PMC7042727
- DOI: 10.1038/s41380-018-0091-8
A rare loss-of-function variant of ADAM17 is associated with late-onset familial Alzheimer disease
Abstract
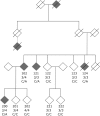
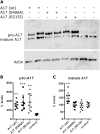
Common variants of about 20 genes contributing to AD risk have so far been identified through genome-wide association studies (GWAS). However, there is still a large proportion of heritability that might be explained by rare but functionally important variants. One of the so far identified genes with rare AD causing variants is ADAM10. Using whole-genome sequencing we now identified a single rare nonsynonymous variant (SNV) rs142946965 [p.R215I] in ADAM17 co-segregating with an autosomal-dominant pattern of late-onset AD in one family. Subsequent genotyping and analysis of available whole-exome sequencing data of additional case/control samples from Germany, UK, and USA identified five variant carriers among AD patients only. The mutation inhibits pro-protein cleavage and the formation of the active enzyme, thus leading to loss-of-function of ADAM17 alpha-secretase. Further, we identified a strong negative correlation between ADAM17 and APP gene expression in human brain and present in vitro evidence that ADAM17 negatively controls the expression of APP. As a consequence, p.R215I mutation of ADAM17 leads to elevated Aß formation in vitro. Together our data supports a causative association of the identified ADAM17 variant in the pathogenesis of AD.
Conflict of interest statement
TG has reported in having received consulting fees from Actelion, Eli Lilly, MSD; Novartis, Quintiles, Roche Pharma, and lecture fees from Biogen, Lilly, Parexel, Roche Pharma, and also grants to his institution from Actelion and PreDemTech. The remaining authors declare that they have no conflict of interest.
Figures


References
-
- Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011;377:1019–31. - PubMed
-
- Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–68. - PubMed
-
- Zunke F, Rose-John S. The shedding protease ADAM17: physiology and pathophysiology. Biochim Biophys Acta. 2017;1864:2059–70. - PubMed
-
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–33. - PubMed
-
- Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, et al. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem. 1998;273:27765–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

