Case-control meta-analysis of blood DNA methylation and autism spectrum disorder
- PMID: 29988321
- PMCID: PMC6022498
- DOI: 10.1186/s13229-018-0224-6
Case-control meta-analysis of blood DNA methylation and autism spectrum disorder
Abstract
Background: Several reports have suggested a role for epigenetic mechanisms in ASD etiology. Epigenome-wide association studies (EWAS) in autism spectrum disorder (ASD) may shed light on particular biological mechanisms. However, studies of ASD cases versus controls have been limited by post-mortem timing and severely small sample sizes. Reports from in-life sampling of blood or saliva have also been very limited in sample size and/or genomic coverage. We present the largest case-control EWAS for ASD to date, combining data from population-based case-control and case-sibling pair studies.
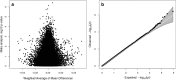
Methods: DNA from 968 blood samples from children in the Study to Explore Early Development (SEED 1) was used to generate epigenome-wide array DNA methylation (DNAm) data at 485,512 CpG sites for 453 cases and 515 controls, using the Illumina 450K Beadchip. The Simons Simplex Collection (SSC) provided 450K array DNAm data on an additional 343 cases and their unaffected siblings. We performed EWAS meta-analysis across results from the two data sets, with adjustment for sex and surrogate variables that reflect major sources of biological variation and technical confounding such as cell type, batch, and ancestry. We compared top EWAS results to those from a previous brain-based analysis. We also tested for enrichment of ASD EWAS CpGs for being targets of meQTL associations using available SNP genotype data in the SEED sample.
Findings: In this meta-analysis of blood-based DNA from 796 cases and 858 controls, no single CpG met a Bonferroni discovery threshold of p < 1.12 × 10- 7. Seven CpGs showed differences at p < 1 × 10- 5 and 48 at 1 × 10- 4. Of the top 7, 5 showed brain-based ASD associations as well, often with larger effect sizes, and the top 48 overall showed modest concordance (r = 0.31) in direction of effect with cerebellum samples. Finally, we observed suggestive evidence for enrichment of CpG sites controlled by SNPs (meQTL targets) among the EWAS CpG hits, which was consistent across EWAS and meQTL discovery p value thresholds.
Conclusions: No single CpG site showed a large enough DNAm difference between cases and controls to achieve epigenome-wide significance in this sample size. However, our results suggest the potential to observe disease associations from blood-based samples. Among the seven sites achieving suggestive statistical significance, we observed consistent, and stronger, effects at the same sites among brain samples. Discovery-oriented EWAS for ASD using blood samples will likely need even larger samples and unified genetic data to further understand DNAm differences in ASD.
Keywords: Autism spectrum disorders; DNA methylation; Epigenome; Peripheral blood; Simons Simplex Collection; Study to Explore Early Development.
Conflict of interest statement
This study was approved by the institutional review boards at each SEED site: SEED 1 recruitment was approved by the IRBs of each recruitment site: Institutional Review Board (IRB)-C, CDC Human Research Protection Office; Kaiser Foundation Research Institute (KFRI) Kaiser Permanente Northern California IRB, Colorado Multiple IRB, Emory University IRB, Georgia Department of Public Health IRB, Maryland Department of Health and Mental Hygiene IRB, Johns Hopkins Bloomberg School of Public Health Review Board, University of North Carolina IRB and Office of Human Research Ethics, IRB of The Children’s Hospital of Philadelphia, and IRB of the University of Pennsylvania. All enrolled families provided written consent for participation. This methylation substudy was approved as an amendment of the Johns Hopkins Institutional Review Board (IRB) approval. For participants from the Simons simplex collection, parents consented and children assented as required by each local institutional review board, which included a coalition of clinics located at Michigan, Yale, Emory, Columbia, Vanderbilt, McGill Washington, and Harvard Universities (Children’s Hospital of Boston), and at the Universities of Washington, Illinois (Chicago), Missouri, UCLA, and the Baylor College of Medicine. To protect the privacy of participants, Global Unique Identifiers (GUIDs) were constructed from personal information using an algorithm devised in collaboration with scientists at the NIH [66]. Each clinic retained personal identifiers on site and transmitted de-identified GUIDs to a central database, as described previously [37].Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

