Detection of Pulmonary Infectious Pathogens From Lung Biopsy Tissues by Metagenomic Next-Generation Sequencing
- PMID: 29988504
- PMCID: PMC6026637
- DOI: 10.3389/fcimb.2018.00205
Detection of Pulmonary Infectious Pathogens From Lung Biopsy Tissues by Metagenomic Next-Generation Sequencing
Abstract
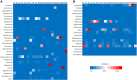
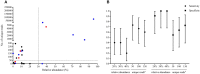
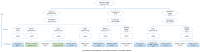
Metagenomic next-generation sequencing (mNGS) is a comprehensive approach for sequence-based identification of pathogenic microbes. However, reports on the use of mNGS in pulmonary infection applied to lung biopsy tissues remain scarce. In this study, we applied mNGS to detect the presence of pathogenic microbes in lung biopsy tissues from 20 patients with pulmonary disorders indicating possible infection. We applied a new data management for identifying pathogen species based on mNGS data. We determined the thresholds for the unique reads and relative abundance required to identify the infectious pathogens. Potential pathogens of pulmonary infections in 15 patients were identified by mNGS. The comparison between mNGS and culture method resulted that the sensitivity and specificity were 100.0% (95% CI: 31.0-100.0%) and 76.5% (95% CI: 49.8-92.2%) for bacteria, 57.1% (95% CI: 20.2-88.2%) and 61.5% (95% CI: 32.2-84.9%) for fungi. The positive predictive value (PPV) (42.9% for bacteria, 44.4% for fungi) was much lower than negative predictive value (NPV) (100% for bacteria, 72.7% for fungi) in mNGS vs. culture method. The mNGS showed the highest specificity (100.0 and 94.1%) and PPV (100.0 and 75.0%) in the evaluation of fungi and MTBC respectively, when compared with histopathology method. The study indicated that mNGS of lung biopsy tissues can be used to detect the presence (or absence) of pulmonary pathogens in patients, with potential benefits in speed and sensitivity. However, accurate data management and interpretation of mNGS are required, and should be combined with observations of clinical manifestations and conventional laboratory-based diagnostic methods.
Keywords: clinical diagnosis; data management; lung biopsy tissues; metagenomic next-generation sequencing (mNGS); pulmonary infection.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

