Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis
- PMID: 29989977
- PMCID: PMC6043423
- DOI: 10.1109/JBHI.2017.2767063
Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis
Abstract
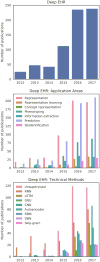
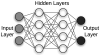
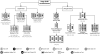
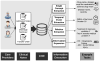
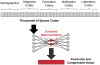
The past decade has seen an explosion in the amount of digital information stored in electronic health records (EHRs). While primarily designed for archiving patient information and performing administrative healthcare tasks like billing, many researchers have found secondary use of these records for various clinical informatics applications. Over the same period, the machine learning community has seen widespread advances in the field of deep learning. In this review, we survey the current research on applying deep learning to clinical tasks based on EHR data, where we find a variety of deep learning techniques and frameworks being applied to several types of clinical applications including information extraction, representation learning, outcome prediction, phenotyping, and deidentification. We identify several limitations of current research involving topics such as model interpretability, data heterogeneity, and lack of universal benchmarks. We conclude by summarizing the state of the field and identifying avenues of future deep EHR research.
Figures

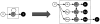

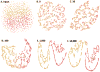

References
-
- Birkhead GS, Klompas M, Shah NR. Uses of Electronic Health Records for Public Health Surveillance to Advance Public Health. Annu Rev Public Health. 2015;36:345–59. - PubMed
-
- T. O. of the National Coordinator for Health Information Technology. Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008-2015, year = 2016. URL = https://dashboard.healthit.gov/evaluations/data-briefs/non-federal-acute....
-
- J E, N N. Electronic Health Record Adoption and Use among Office-based Physicians in the U.S., by State: 2015 National Electronic Health Records Survey. The Office of the National Coordinator for Health Information Technology, Tech Rep. 2016
-
- Botsis T, Hartvigsen G, Chen F, Weng C. Secondary Use of ehr: Data Quality Issues and Informatics Opportunities. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2010;2010:1–5. [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/21347133{%}5Cnhttp://www.pubmedcentra.... - PMC - PubMed
-
- Jensen PB, Jensen LJ, Brunak S. Translational genetics: Mining electronic health records: towards better research applications and clinical care. Nature Reviews - Genetics. 2012;13:395–405. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

