Arabidopsis Leaf Flatness Is Regulated by PPD2 and NINJA through Repression of CYCLIN D3 Genes
- PMID: 29991485
- PMCID: PMC6130026
- DOI: 10.1104/pp.18.00327
Arabidopsis Leaf Flatness Is Regulated by PPD2 and NINJA through Repression of CYCLIN D3 Genes
Abstract
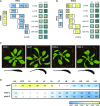
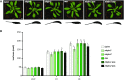
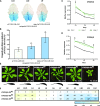
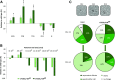
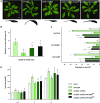
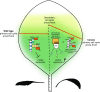
In Arabidopsis (Arabidopsis thaliana), reduced expression of the transcriptional regulator PEAPOD2 (PPD2) results in propeller-like rosettes with enlarged and dome-shaped leaves. However, the molecular and cellular processes underlying this peculiar phenotype remain elusive. Here, we studied the interaction between PPD2 and NOVEL INTERACTOR OF JAZ (NINJA) and demonstrated that ninja loss-of-function plants produce rosettes with dome-shaped leaves similar to those of ppd mutants but without the increase in size. We showed that ninja mutants have a convex-shaped primary cell cycle arrest front, putatively leading to excessive cell division in the central leaf blade region. Furthermore, ppd and ninja mutants have a similar increase in the expression of CYCLIN D3;2 (CYCD3;2), and ectopic overexpression of CYCD3;2 phenocopies the ppd and ninja rosette and leaf shape phenotypes without affecting the size. Our results reveal a pivotal contribution of NINJA in leaf development, in addition to its well-studied function in jasmonate signaling, and imply a new function for D3-type cyclins in, at least partially, uncoupling the size and shape phenotypes of ppd leaves.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 - PubMed
-
- Bai Y, Meng Y, Huang D, Qi Y, Chen M (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98: 128–136 - PubMed
-
- Bergmann DC, Sack FD (2007) Stomatal development. Annu Rev Plant Biol 58: 163–181 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

