Evolutionary history of human colitis-associated colorectal cancer
- PMID: 29991641
- PMCID: PMC6580738
- DOI: 10.1136/gutjnl-2018-316191
Evolutionary history of human colitis-associated colorectal cancer
Abstract
Objective: IBD confers an increased lifetime risk of developing colorectal cancer (CRC), and colitis-associated CRC (CA-CRC) is molecularly distinct from sporadic CRC (S-CRC). Here we have dissected the evolutionary history of CA-CRC using multiregion sequencing.
Design: Exome sequencing was performed on fresh-frozen multiple regions of carcinoma, adjacent non-cancerous mucosa and blood from 12 patients with CA-CRC (n=55 exomes), and key variants were validated with orthogonal methods. Genome-wide copy number profiling was performed using single nucleotide polymorphism arrays and low-pass whole genome sequencing on archival non-dysplastic mucosa (n=9), low-grade dysplasia (LGD; n=30), high-grade dysplasia (HGD; n=13), mixed LGD/HGD (n=7) and CA-CRC (n=19). Phylogenetic trees were reconstructed, and evolutionary analysis used to reveal the temporal sequence of events leading to CA-CRC.
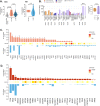
Results: 10/12 tumours were microsatellite stable with a median mutation burden of 3.0 single nucleotide alterations (SNA) per Mb, ~20% higher than S-CRC (2.5 SNAs/Mb), and consistent with elevated ageing-associated mutational processes. Non-dysplastic mucosa had considerable mutation burden (median 47 SNAs), including mutations shared with the neighbouring CA-CRC, indicating a precancer mutational field. CA-CRCs were often near triploid (40%) or near tetraploid (20%) and phylogenetic analysis revealed that copy number alterations (CNAs) began to accrue in non-dysplastic bowel, but the LGD/HGD transition often involved a punctuated 'catastrophic' CNA increase.
Conclusions: Evolutionary genomic analysis revealed precancer clones bearing extensive SNAs and CNAs, with progression to cancer involving a dramatic accrual of CNAs at HGD. Detection of the cancerised field is an encouraging prospect for surveillance, but punctuated evolution may limit the window for early detection.
Keywords: IBD - genetics; colorectal cancer; dysplasia; inflammatory bowel disease.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
Publication types
MeSH terms
Grants and funding
- A16581/CRUK_/Cancer Research UK/United Kingdom
- A19771/CRUK_/Cancer Research UK/United Kingdom
- 16459/CRUK_/Cancer Research UK/United Kingdom
- MR/L016508/1/MRC_/Medical Research Council/United Kingdom
- 14895/CRUK_/Cancer Research UK/United Kingdom
- MR/P00122X/1/MRC_/Medical Research Council/United Kingdom
- MR/S003851/1/MRC_/Medical Research Council/United Kingdom
- 19771/CRUK_/Cancer Research UK/United Kingdom
- 206314/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- 25901/CRUK_/Cancer Research UK/United Kingdom
- A14895/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
