Integrative genomic analysis of adult mixed phenotype acute leukemia delineates lineage associated molecular subtypes
- PMID: 29991687
- PMCID: PMC6039465
- DOI: 10.1038/s41467-018-04924-z
Integrative genomic analysis of adult mixed phenotype acute leukemia delineates lineage associated molecular subtypes
Abstract
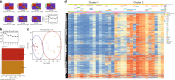
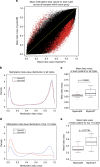
Mixed phenotype acute leukemia (MPAL) is a rare subtype of acute leukemia characterized by leukemic blasts presenting myeloid and lymphoid markers. Here we report data from integrated genomic analysis on 31 MPAL samples and compare molecular profiling with that from acute myeloid leukemia (AML), B cell acute lymphoblastic leukemia (B-ALL), and T cell acute lymphoblastic leukemia (T-ALL). Consistent with the mixed immunophenotype, both AML-type and ALL-type mutations are detected in MPAL. Myeloid-B and myeloid-T MPAL show distinct mutation and methylation signatures that are associated with differences in lineage-commitment gene expressions. Genome-wide methylation comparison among MPAL, AML, B-ALL, and T-ALL sub-classifies MPAL into AML-type and ALL-type MPAL, which is associated with better clinical response when lineage-matched therapy is given. These results elucidate the genetic and epigenetic heterogeneity of MPAL and its genetic distinction from AML, B-ALL, and T-ALL and further provide proof of concept for a molecularly guided precision therapy approach in MPAL.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

