Cross-feeding modulates antibiotic tolerance in bacterial communities
- PMID: 29991761
- PMCID: PMC6194032
- DOI: 10.1038/s41396-018-0212-z
Cross-feeding modulates antibiotic tolerance in bacterial communities
Abstract
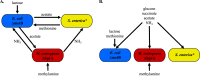
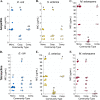
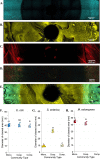
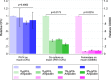
Microbes frequently rely on metabolites excreted by other bacterial species, but little is known about how this cross-feeding influences the effect of antibiotics. We hypothesized that when species rely on each other for essential metabolites, the minimum inhibitory concentration (MIC) for all species will drop to that of the "weakest link"-the species least resistant in monoculture. We tested this hypothesis in an obligate cross-feeding system that was engineered between Escherichia coli, Salmonella enterica, and Methylobacterium extorquens. The effect of tetracycline and ampicillin were tested on both liquid and solid media. In all cases, resistant species were inhibited at significantly lower antibiotic concentrations in the cross-feeding community than in monoculture or a competitive community. However, deviation from the "weakest link" hypothesis was also observed in cross-feeding communities apparently as result of changes in the timing of growth and cross-protection. Comparable results were also observed in a clinically relevant system involving facultative cross-feeding between Pseudomonas aeruginosa and an anaerobic consortium found in the lungs of cystic fibrosis patients. P. aeruginosa was inhibited by lower concentrations of ampicillin when cross-feeding than when grown in isolation. These results suggest that cross-feeding significantly alters tolerance to antibiotics in a variety of systems.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva: WHO; 2014.
-
- Friman V.-P., Guzman L. M., Reuman D. C., Bell T. Bacterial adaptation to sublethal antibiotic gradients can change the ecological properties of multitrophic microbial communities. Proceedings of the Royal Society B: Biological Sciences. 2015;282(1806):20142920–20142920. doi: 10.1098/rspb.2014.2920. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

