Promoter methylation of the MGAT3 and BACH2 genes correlates with the composition of the immunoglobulin G glycome in inflammatory bowel disease
- PMID: 29991969
- PMCID: PMC5987481
- DOI: 10.1186/s13148-018-0507-y
Promoter methylation of the MGAT3 and BACH2 genes correlates with the composition of the immunoglobulin G glycome in inflammatory bowel disease
Abstract
Background: Many genome- and epigenome-wide association studies (GWAS and EWAS) and studies of promoter methylation of candidate genes for inflammatory bowel disease (IBD) have demonstrated significant associations between genetic and epigenetic changes and IBD. Independent GWA studies have identified genetic variants in the BACH2, IL6ST, LAMB1, IKZF1, and MGAT3 loci to be associated with both IBD and immunoglobulin G (IgG) glycosylation.
Methods: Using bisulfite pyrosequencing, we analyzed CpG methylation in promoter regions of these five genes from peripheral blood of several hundred IBD patients and healthy controls (HCs) from two independent cohorts, respectively.
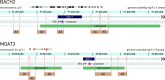
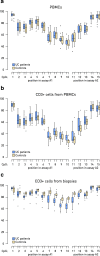
Results: We found significant differences in the methylation levels in the MGAT3 and BACH2 genes between both Crohn's disease and ulcerative colitis when compared to HC. The same pattern of methylation changes was identified for both genes in CD19+ B cells isolated from the whole blood of a subset of the IBD patients. A correlation analysis was performed between the MGAT3 and BACH2 promoter methylation and individual IgG glycans, measured in the same individuals of the two large cohorts. MGAT3 promoter methylation correlated significantly with galactosylation, sialylation, and bisecting GlcNAc on IgG of the same patients, suggesting that activity of the GnT-III enzyme, encoded by this gene, might be altered in IBD. The correlations between the BACH2 promoter methylation and IgG glycans were less obvious, since BACH2 is not a glycosyltransferase and therefore may affect IgG glycosylation only indirectly.
Conclusions: Our results suggest that epigenetic deregulation of key glycosylation genes might lead to an increase in pro-inflammatory properties of IgG in IBD through a decrease in galactosylation and sialylation and an increase of bisecting GlcNAc on digalactosylated glycan structures. Finally, we showed that CpG methylation in the promoter of the MGAT3 gene is altered in CD3+ T cells isolated from inflamed mucosa of patients with ulcerative colitis from a third smaller cohort, for which biopsies were available, suggesting a functional role of this glyco-gene in IBD pathogenesis.
Conflict of interest statement
For the Edinburgh and Florence cohorts, ethical approvals were obtained from Tayside Committee on Medical Ethics B, and all patients and controls provided written, informed consent (LREC 06/S1101/16, LREC 2000/4/192). For the patients from Portugal, all clinical procedures and research protocols were approved by the ethics committee of CHP/HSA, Portugal (233/12(179-DEFI/177-CES); all participants gave their informed consent.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Burisch J, Pedersen N, Čuković-Čavka S, Brinar M, Kaimakliotis I, Duricova D, Shonová O, Vind I, Avnstrøm S, Thorsgaard N, et al. East–West gradient in the incidence of inflammatory bowel disease in Europe: the ECCO-EpiCom inception cohort. Gut. 2013;63:588–597. doi: 10.1136/gutjnl-2013-304636. - DOI - PubMed
-
- Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

