Whole-genome sequencing reveals highly specific gene targeting by in vitro assembled Cas9-ribonucleoprotein complexes in Aspergillus fumigatus
- PMID: 29992034
- PMCID: PMC5987418
- DOI: 10.1186/s40694-018-0057-2
Whole-genome sequencing reveals highly specific gene targeting by in vitro assembled Cas9-ribonucleoprotein complexes in Aspergillus fumigatus
Abstract
Background: CRISPR/Cas9-based genome editing is quickly becoming a powerful tool within the field of fungal genetics. Adaptation of CRISPR/Cas9 systems are allowing for rapid and highly efficient gene targeting within fungi. We recently reported the adaptation of a simple CRISPR/Cas9 system for gene deletion that is effective across multiple genetic backgrounds of Aspergillus fumigatus. This system employs in vitro assembly of Cas9 ribonucleoproteins (RNPs) coupled with micro-homology repair templates for gene deletion. Although highly efficient at gene targeting in wild type genetic backgrounds of A. fumigatus, the potential for our system to produce unwanted off-target mutations has not been addressed.
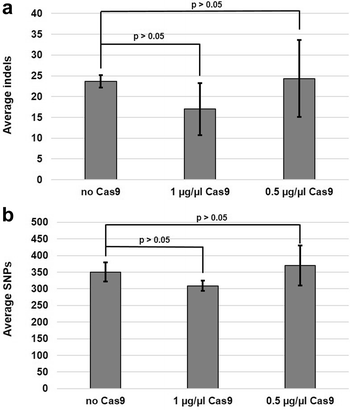
Results: Next-generation Illumina sequencing was used to identify genome mutations among transformants isolated from standard (no Cas9) and Cas9-mediated integration of a hygromycin deletion cassette. Two different concentrations of Cas9 were utilized to examine the association of Cas9 concentration with total numbers and types of genomic mutations. For each of the three test groups (zero, low, and high Cas9), three transformants were sequenced and compared to the parent strain. Bioinformatics analyses revealed the average number of total mutations to be similar among all three test groups. A. fumigatus transformation using standard, non-Cas9-mediated methods resulted in an average of 373 ± 28 mutations. In comparison, transformation with in vitro assembled Cas9-RNPs using either high (1 µg/µl) or low (0.5 µg/µl) levels of Cas9 resulted in an average of 326 ± 19 and 395 ± 69 mutations, respectively. In all cases, the vast majority of mutations identified were intergenic. No correlation between the amount of Cas9 utilized for transformation and the overall number of mutations was found. Finally, the specific type of mutation introduced during the transformation process was not Cas9-dependent, as both single-nucleotide polymorphisms and insertion/deletion events were not significantly different between the experimental groups.
Conclusions: CRISPR/Cas9-based genome editing in A. fumigatus using in vitro assembled RNPs coupled with microhomology templates is a reliable method of gene targeting. This system is highly efficient and is not associated with increased off-target mutations caused by introduction of the Cas9 nuclease.
Keywords: Aspergillus fumigatus; CRISPR/Cas9; Genome editing; Off-target mutation.
Figures
References
-
- da Silva Ferreira ME, Kress MRVZ, Savoldi M, Goldman MHS, Härtl A, Heinekamp T, Brakhage AA, Goldman GH. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

