Reference Gene Selection for Quantitative Real-Time PCR of Mycelia from Lentinula edodes under High-Temperature Stress
- PMID: 29992134
- PMCID: PMC6016149
- DOI: 10.1155/2018/1670328
Reference Gene Selection for Quantitative Real-Time PCR of Mycelia from Lentinula edodes under High-Temperature Stress
Abstract
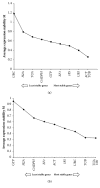
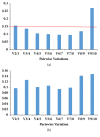
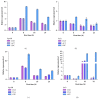
Housekeeping genes are important for measuring the transcription expression of functional genes; 10 traditional reference genes, TUB, TUA, GADPH, EF1, 18S, GTP, ACT, UBI, UBC, and H2A, were tested for their adequacy in Lentinula edodes (L. edodes). Using specific primers, mRNA levels of these candidate housekeeping genes were evaluated in mycelia of L. edodes, which were treated with high-temperature stress at 37°C for 0, 4, 8, 12, 18, and 24 hours. After treatment, expression stability of candidate genes was evaluated using three statistical software programs: geNorm, NormFinder, and BestKeeper. According to geNorm, TUB had the lowest M values in L. edodes strains 18 and 18N44. Using NormFinder, the best candidate reference gene in strain 18 was TUB (0.030), and the best candidate reference gene in strain 18N44 was UBI (0.047). In BestKeeper analysis, the standard deviation (SD) values of UBC, TUA, H2A, EF1, ACT, 18S, and GTP in strain 18 and those of GADPH and GTP in strain 18N44 were greater than 1; thus, these genes were disqualified as reference genes. Taken together, only UBI and TUB were found to be desirable reference genes by BestKeeper software. Based on the results of three software analyses, TUB was the most stable gene under all conditions and was verified as an appropriate reference gene for quantitative real-time polymerase chain reaction in L. edodes mycelia under high-temperature stress.
Figures


Similar articles
-
Selection and Validation of Reference Genes for qRT-PCR in Lentinula edodes under Different Experimental Conditions.Genes (Basel). 2019 Aug 27;10(9):647. doi: 10.3390/genes10090647. Genes (Basel). 2019. PMID: 31461882 Free PMC article.
-
Identification and evaluation of reference genes for qRT-PCR studies in Lentinula edodes.PLoS One. 2018 Jan 2;13(1):e0190226. doi: 10.1371/journal.pone.0190226. eCollection 2018. PLoS One. 2018. PMID: 29293626 Free PMC article.
-
Selection of Reference Genes for qRT-PCR Analysis in Lentinula edodes after Hot-Air Drying.Molecules. 2018 Dec 31;24(1):136. doi: 10.3390/molecules24010136. Molecules. 2018. PMID: 30602709 Free PMC article.
-
[Selection and validation of reference genes for quantitative Real-time PCR analysis in Amana edulis].Zhongguo Zhong Yao Za Zhi. 2021 Feb;46(4):938-943. doi: 10.19540/j.cnki.cjcmm.20201120.104. Zhongguo Zhong Yao Za Zhi. 2021. PMID: 33645100 Chinese.
-
GC⁻MS-Based Nontargeted and Targeted Metabolic Profiling Identifies Changes in the Lentinula edodes Mycelial Metabolome under High-Temperature Stress.Int J Mol Sci. 2019 May 10;20(9):2330. doi: 10.3390/ijms20092330. Int J Mol Sci. 2019. PMID: 31083449 Free PMC article.
Cited by
-
Selection and validation of reference genes for RT-qPCR in ophiocordyceps sinensis under different experimental conditions.PLoS One. 2024 Feb 6;19(2):e0287882. doi: 10.1371/journal.pone.0287882. eCollection 2024. PLoS One. 2024. PMID: 38319940 Free PMC article.
-
Relative quantification of BCL2 mRNA for diagnostic usage needs stable uncontrolled genes as reference.PLoS One. 2020 Aug 12;15(8):e0236338. doi: 10.1371/journal.pone.0236338. eCollection 2020. PLoS One. 2020. PMID: 32785215 Free PMC article.
-
Heat stress-induced NO enhanced perylenequinone biosynthesis of Shiraia sp. via calcium signaling pathway.Appl Microbiol Biotechnol. 2024 May 3;108(1):317. doi: 10.1007/s00253-024-13142-1. Appl Microbiol Biotechnol. 2024. PMID: 38700737 Free PMC article.
-
Genome-Wide Screening and Stability Verification of the Robust Internal Control Genes for RT-qPCR in Filamentous Fungi.J Fungi (Basel). 2022 Sep 10;8(9):952. doi: 10.3390/jof8090952. J Fungi (Basel). 2022. PMID: 36135677 Free PMC article.
-
Screening and validating of endogenous reference genes in Chlorella sp. TLD 6B under abiotic stress.Sci Rep. 2023 Jan 27;13(1):1555. doi: 10.1038/s41598-023-28311-x. Sci Rep. 2023. PMID: 36707665 Free PMC article.
References
-
- Goulao L. F., Fortunato A. S., Ramalho J. C. Selection of Reference Genes for Normalizing Quantitative Real-Time PCR Gene Expression Data with Multiple Variables in Coffea spp. Plant Molecular Biology Reporter. 2012;30(3):741–759. doi: 10.1007/s11105-011-0382-6. - DOI
-
- Pfaffl M. Quantification Strategies in Real-time Polymerase Chain Reaction. 2012.
-
- Dong E. N., Liang Q., Li L., et al. The selection of reference gene in real-time quantitative reverse transcription PCR. Chinese Journal of Animal Science. 2013;49(11):92–96.
-
- Huang Z. C., Ouyang L. J., Zhang L., Sha Y. E., Zeng F. H. Selection and evaluation of internal reference genes in Eucalyptus species. Journal of Northwest AF University (Nat. Sci) 2013;41(10):67–72.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

