Reference Gene Selection for Quantitative Real-Time PCR of Mycelia from Lentinula edodes under High-Temperature Stress
- PMID: 29992134
- PMCID: PMC6016149
- DOI: 10.1155/2018/1670328
Reference Gene Selection for Quantitative Real-Time PCR of Mycelia from Lentinula edodes under High-Temperature Stress
Abstract
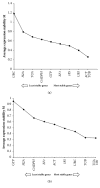
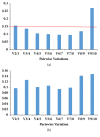
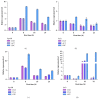
Housekeeping genes are important for measuring the transcription expression of functional genes; 10 traditional reference genes, TUB, TUA, GADPH, EF1, 18S, GTP, ACT, UBI, UBC, and H2A, were tested for their adequacy in Lentinula edodes (L. edodes). Using specific primers, mRNA levels of these candidate housekeeping genes were evaluated in mycelia of L. edodes, which were treated with high-temperature stress at 37°C for 0, 4, 8, 12, 18, and 24 hours. After treatment, expression stability of candidate genes was evaluated using three statistical software programs: geNorm, NormFinder, and BestKeeper. According to geNorm, TUB had the lowest M values in L. edodes strains 18 and 18N44. Using NormFinder, the best candidate reference gene in strain 18 was TUB (0.030), and the best candidate reference gene in strain 18N44 was UBI (0.047). In BestKeeper analysis, the standard deviation (SD) values of UBC, TUA, H2A, EF1, ACT, 18S, and GTP in strain 18 and those of GADPH and GTP in strain 18N44 were greater than 1; thus, these genes were disqualified as reference genes. Taken together, only UBI and TUB were found to be desirable reference genes by BestKeeper software. Based on the results of three software analyses, TUB was the most stable gene under all conditions and was verified as an appropriate reference gene for quantitative real-time polymerase chain reaction in L. edodes mycelia under high-temperature stress.
Figures


References
-
- Goulao L. F., Fortunato A. S., Ramalho J. C. Selection of Reference Genes for Normalizing Quantitative Real-Time PCR Gene Expression Data with Multiple Variables in Coffea spp. Plant Molecular Biology Reporter. 2012;30(3):741–759. doi: 10.1007/s11105-011-0382-6. - DOI
-
- Pfaffl M. Quantification Strategies in Real-time Polymerase Chain Reaction. 2012.
-
- Dong E. N., Liang Q., Li L., et al. The selection of reference gene in real-time quantitative reverse transcription PCR. Chinese Journal of Animal Science. 2013;49(11):92–96.
-
- Huang Z. C., Ouyang L. J., Zhang L., Sha Y. E., Zeng F. H. Selection and evaluation of internal reference genes in Eucalyptus species. Journal of Northwest AF University (Nat. Sci) 2013;41(10):67–72.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

