Mechanism of BRAF Activation through Biochemical Characterization of the Recombinant Full-Length Protein
- PMID: 29992710
- PMCID: PMC6504641
- DOI: 10.1002/cbic.201800359
Mechanism of BRAF Activation through Biochemical Characterization of the Recombinant Full-Length Protein
Abstract
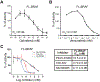
BRAF kinase plays an important role in mitogen-activated protein kinase (MAPK) signaling and harbors activating mutations in about half of melanomas and in a smaller percentage in many other cancers. Despite its importance, few in vitro studies have been performed to characterize the biochemical properties of full-length BRAF. Herein, a strategy to generate an active, intact form of BRAF protein suitable for in vitro enzyme kinetics is described. It is shown that purified, intact BRAF protein autophosphorylates the kinase activation loop and this can be enhanced by binding the MEK protein substrate through an allosteric mechanism. These studies provide in vitro evidence that BRAF selectively binds to active RAS and that the BRAF/CRAF heterodimer is the most active form, relative to their respective homodimers. Full-length BRAF analysis with small-molecule BRAF inhibitors shows that two drugs, dabrafenib and vemurafenib, can modestly enhance kinase activity of BRAF at low concentration. Taken together, this characterization of intact BRAF contributes to a framework for understanding its role in cell signaling.
Keywords: BRAF; cancer; cell signaling; enzymes; proteins; reaction mechanisms.
© 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures




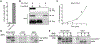
References
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA, Nature 2002, 417, 949–954 - PubMed
- Holderfield M, Deuker MM, McCormick F, McMahon M, Nat Rev Cancer 2014, 14, 455–467. - PMC - PubMed
-
- Lavoie H, Therrien M, Nat Rev Mol Cell Biol 2015, 16, 281–298. - PubMed
-
- Muslin AJ, Tanner JW, Allen PM, Shaw AS, Cell 1996, 84, 889–897. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

