Small Regulatory RNAs in the Enterobacterial Response to Envelope Damage and Oxidative Stress
- PMID: 29992897
- PMCID: PMC10361636
- DOI: 10.1128/microbiolspec.RWR-0022-2018
Small Regulatory RNAs in the Enterobacterial Response to Envelope Damage and Oxidative Stress
Abstract
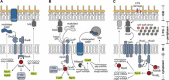
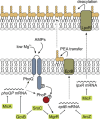
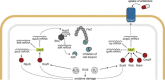
The ability of bacteria to thrive in diverse habitats and to adapt to ever-changing environmental conditions relies on the rapid and stringent modulation of gene expression. It has become evident in the past decade that small regulatory RNAs (sRNAs) are central components of networks controlling the bacterial responses to stress. Functioning at the posttranscriptional level, sRNAs base-pair with cognate mRNAs to alter translation, stability, or both to either repress or activate the targeted transcripts; the RNA chaperone Hfq participates in stabilizing sRNAs and in promoting pairing between target and sRNA. In particular, sRNAs act at the heart of crucial stress responses, including those dedicated to overcoming membrane damage and oxidative stress, discussed here. The bacterial cell envelope is the outermost protective barrier against the environment and thus is constantly monitored and remodeled. Here, we review the integration of sRNAs into the complex networks of several major envelope stress responses of Gram-negative bacteria, including the RpoE (σE), Cpx, and Rcs regulons. Oxidative stress, caused by bacterial respiratory activity or induced by toxic molecules, can lead to significant damage of cellular components. In Escherichia coli and related bacteria, sRNAs also contribute significantly to the function of the RpoS (σS)-dependent general stress response as well as the specific OxyR- and SoxR/S-mediated responses to oxidative damage. Their activities in gene regulation and crosstalk to other stress-induced regulons are highlighted.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

