The influenza virus hemagglutinin head evolves faster than the stalk domain
- PMID: 29992986
- PMCID: PMC6041311
- DOI: 10.1038/s41598-018-28706-1
The influenza virus hemagglutinin head evolves faster than the stalk domain
Abstract
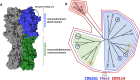
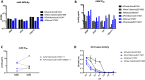
The limited ability of current influenza virus vaccines to protect from antigenically drifted or shifted viruses creates a public health problem that has led to the need to develop effective, broadly protective vaccines. While current influenza virus vaccines mostly induce an immune response against the immunodominant and variable head domain of the hemagglutinin, the major surface glycoprotein of the virus, the hemagglutinin stalk domain has been identified to harbor neutralizing B-cell epitopes that are conserved among and even between influenza A virus subtypes. A complete understanding of the differences in evolution between the main target of current vaccines and this more conserved stalk region are missing. Here, we performed an evolutionary analysis of the stalk domains of the hemagglutinin of pre-pandemic seasonal H1N1, pandemic H1N1, seasonal H3N2, and influenza B viruses and show quantitatively for the first time that the stalk domain is evolving at a rate that is significantly slower than that of the head domain. Additionally, we found that the cross-reactive epitopes in the stalk domain targeted by broadly neutralizing monoclonal antibodies are evolving at an even slower rate compared to the full head and stalk regions of the protein. Finally, a fixed-effects likelihood selection analysis was performed for these virus groups in both the head and stalk domains. While several positive selection sites were found in the head domain, only a single site in the stalk domain of pre-pandemic seasonal H1 hemagglutinin was identified at amino acid position 468 (H1 numbering from methionine). This site is not located in or close to the epitopes of cross-reactive anti-stalk monoclonal antibodies. Furthermore, we found that changes in this site do not significantly impact virus binding or neutralization by human anti-stalk antibodies, suggesting that some positive selection in the stalk domain is independent of immune pressures. We conclude that, while the stalk domain does evolve over time, this evolution is slow and, historically, is not directed to aid in evading neutralizing antibody responses.
Conflict of interest statement
The Icahn School of Medicine at Mount Sinai has filed patents regarding influenza virus vaccines.
Figures



References
-
- Cox, N. & Subbarao, K. Global epidemiology of influenza: past and present. Annu Rev Med51, 10.1146/annurev.med.51.1.407 (2000). - PubMed
-
- Office of the Associate Director for Communication, D. M. B., Division of Public Affairs. Seasonal Influenza Vaccine Effectiveness, 2005–2016 (2016).
-
- Shaw, M. & Palese, P. (eds Howley, P. M. & Knipe, D. M.) Ch. Orthomyxoviridae, 1151–1185 (Lippincott-Raven, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

