The composition of the perinatal intestinal microbiota in cattle
- PMID: 29993024
- PMCID: PMC6041309
- DOI: 10.1038/s41598-018-28733-y
The composition of the perinatal intestinal microbiota in cattle
Erratum in
-
Publisher Correction: The composition of the perinatal intestinal microbiota in cattle.Sci Rep. 2018 Sep 11;8(1):13792. doi: 10.1038/s41598-018-31494-3. Sci Rep. 2018. PMID: 30206238 Free PMC article.
Abstract
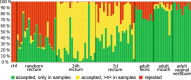
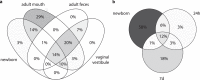
Recent research suggests that the microbial colonization of the mammalian intestine may begin before birth, but the observations are controversial due to challenges in the reliable sampling and analysis of low-abundance microbiota. We studied the perinatal microbiota of calves by sampling them immediately at birth and during the first postnatal week. The large size of the bovine newborns allows sampling directly from rectum using contamination-shielded swabs. Our 16S rDNA data, purged of potential contaminant sequences shared with negative controls, indicates the existence of a diverse low-abundance microbiota in the newborn rectal meconium and mucosa. The newborn rectal microbiota was composed of Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes. The microbial profile resembled dam oral rather than fecal or vaginal vestibular microbiota, but included typical intestinal taxa. During the first postnatal day, the rectum was invaded by Escherichia/Shigella and Clostridia, and the diversity collapsed. By 7 days, diversity was again increasing. In terms of relative abundance, Proteobacteria were replaced by Firmicutes, Bacteroidetes and Actinobacteria, including Faecalibacterium, Bacteroides, Lactobacillus, Butyricicoccus and Bifidobacterium. Our observations suggest that mammals are seeded before birth with a diverse microbiota, but the microbiota changes rapidly in the early postnatal life.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

