Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes
- PMID: 29995839
- PMCID: PMC6461848
- DOI: 10.1038/s41396-018-0175-0
Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes
Abstract
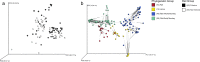
Over the past decade several studies have reported that the gut microbiomes of mammals with similar dietary niches exhibit similar compositional and functional traits. However, these studies rely heavily on samples from captive individuals and often confound host phylogeny, gut morphology, and diet. To more explicitly test the influence of host dietary niche on the mammalian gut microbiome we use 16S rRNA gene amplicon sequencing and shotgun metagenomics to compare the gut microbiota of 18 species of wild non-human primates classified as either folivores or closely related non-folivores, evenly distributed throughout the primate order and representing a range of gut morphological specializations. While folivory results in some convergent microbial traits, collectively we show that the influence of host phylogeny on both gut microbial composition and function is much stronger than that of host dietary niche. This pattern does not result from differences in host geographic location or actual dietary intake at the time of sampling, but instead appears to result from differences in host physiology. These findings indicate that mammalian gut microbiome plasticity in response to dietary shifts over both the lifespan of an individual host and the evolutionary history of a given host species is constrained by host physiological evolution. Therefore, the gut microbiome cannot be considered separately from host physiology when describing host nutritional strategies and the emergence of host dietary niches.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

