Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA
- PMID: 29996105
- PMCID: PMC6269105
- DOI: 10.1016/j.celrep.2018.06.021
Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA
Abstract
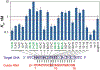
Prokaryotic Argonaute (Ago) proteins were recently shown to target foreign genetic elements, thus making them a perfect model for studies of interference mechanisms. Here, we study interactions of Rhodobacter sphaeroides Ago (RsAgo) with guide RNA (gRNA) and fully complementary or imperfect target DNA (tDNA) using biochemical and structural approaches. We show that RsAgo can specifically recognize both the first nucleotide in gRNA and complementary nucleotide in tDNA, and both interactions contribute to nucleic acid binding. Non-canonical pairs and bulges on the target strand can be accommodated by RsAgo with minimal perturbation of the duplex but significantly reduce RsAgo affinity to tDNA. Surprisingly, mismatches between gRNA and tDNA induce dissociation of the guide-target duplex from RsAgo. Our results reveal plasticity in the ability of Ago proteins to accommodate helical imperfections, show how this might affect the efficiency of RNA silencing, and suggest a potential mechanism for guide release and Ago recycling.
Keywords: RNA-DNA heteroduplex; Rhodobacter sphaeroides Argonaute; RsAgo; guide RNA; non-canonical base pairs and bulges; target DNA.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





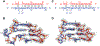
Similar articles
-
Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.Nat Commun. 2016 Jun 21;7:11846. doi: 10.1038/ncomms11846. Nat Commun. 2016. PMID: 27325485 Free PMC article.
-
Recognition of double-stranded DNA by the Rhodobacter sphaeroides Argonaute protein.Biochem Biophys Res Commun. 2020 Dec 17;533(4):1484-1489. doi: 10.1016/j.bbrc.2020.10.051. Epub 2020 Oct 24. Biochem Biophys Res Commun. 2020. PMID: 33333714
-
Bacterial argonaute samples the transcriptome to identify foreign DNA.Mol Cell. 2013 Sep 12;51(5):594-605. doi: 10.1016/j.molcel.2013.08.014. Mol Cell. 2013. PMID: 24034694 Free PMC article.
-
Argonaute proteins: structures and their endonuclease activity.Mol Biol Rep. 2021 May;48(5):4837-4849. doi: 10.1007/s11033-021-06476-w. Epub 2021 Jun 11. Mol Biol Rep. 2021. PMID: 34117606 Review.
-
Ancestral roles of small RNAs: an Ago-centric perspective.Cold Spring Harb Perspect Biol. 2011 Oct 1;3(10):a003772. doi: 10.1101/cshperspect.a003772. Cold Spring Harb Perspect Biol. 2011. PMID: 20810548 Free PMC article. Review.
Cited by
-
In vitro characterization of a pAgo nuclease TtdAgo from Thermococcus thioreducens and evaluation of its effect in vivo.Front Bioeng Biotechnol. 2023 Mar 2;11:1142637. doi: 10.3389/fbioe.2023.1142637. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 36937752 Free PMC article.
-
NAT10 and DDX21 Proteins Interact with RNase H1 and Affect the Performance of Phosphorothioate Oligonucleotides.Nucleic Acid Ther. 2022 Aug;32(4):280-299. doi: 10.1089/nat.2021.0107. Epub 2022 Jul 18. Nucleic Acid Ther. 2022. PMID: 35852833 Free PMC article.
-
An Argonaute from Thermus parvatiensis exhibits endonuclease activity mediated by 5' chemically modified DNA guides.Acta Biochim Biophys Sin (Shanghai). 2022 May 25;54(5):686-695. doi: 10.3724/abbs.2022047. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35643958 Free PMC article.
-
Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea.Nucleic Acids Res. 2019 Jun 20;47(11):5822-5836. doi: 10.1093/nar/gkz379. Nucleic Acids Res. 2019. PMID: 31114878 Free PMC article.
-
Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus.RNA Biol. 2020 May;17(5):677-688. doi: 10.1080/15476286.2020.1724716. Epub 2020 Feb 16. RNA Biol. 2020. PMID: 32013676 Free PMC article.
References
-
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

