Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.)
- PMID: 29996483
- PMCID: PMC6073244
- DOI: 10.3390/ijms19072009
Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.)
Abstract
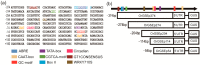
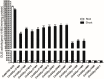
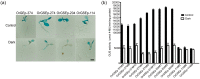
Promoters play a very important role in the initiation and regulation of gene transcription. Green-tissue promoter is of great significance to the development of genetically modified crops. Based on RNA-seq data and RT-PCR expression analysis, this study screened a gene, OrGSE (GREEN SPECIAL EXPRESS), which is expressed specifically in green tissues. The study also isolated the promoter of the OrGSE gene (OrGSEp), and predicted many cis-acting elements, such as the CAAT-Box and TATA-Box, and light-responding elements, including circadian, G-BOX and GT1 CONSENSUS. Histochemical analysis and quantification of GUS activity in transgenic Arabidopsis thaliana plants expressing GUS under the control of OrGSEp revealed that this promoter is not only green tissue-specific, but also light-inducible. The ability of a series of 5’-deletion fragments of OrGSEp to drive GUS expression in Arabidopsis was also evaluated. We found that the promoter region from −54 to −114 is critical for the promoter function, and the region from −374 to −114 may contain core cis-elements involved in light response. In transgenic rice expressing GUS under the control of OrGSEp, visualization and quantification of GUS activity showed that GUS was preferentially expressed in green tissues and not in endosperm. OrGSEp is a useful regulatory element for breeding pest-resistant crops.
Keywords: Green tissue-specific expression; Promoter; common wild rice; light-induced.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Pih K.T., Yoo J., Fosket D.E., Han I.S. A comparison of the activity of three cauliflower mosaic virus 35s promoters in rice seedlings and tobacco (by-2) protoplasts by analysis of gus reporter gene transient expression. Plant. Sci. 1996;114:141–148. doi: 10.1016/0168-9452(95)04316-0. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

