Viral diversity is an obligate consideration in CRISPR/Cas9 designs for targeting the HIV reservoir
- PMID: 29996827
- PMCID: PMC6040082
- DOI: 10.1186/s12915-018-0544-1
Viral diversity is an obligate consideration in CRISPR/Cas9 designs for targeting the HIV reservoir
Abstract
Background: RNA-guided CRISPR/Cas9 systems can be designed to mutate or excise the integrated HIV genome from latently infected cells and have therefore been proposed as a curative approach for HIV. However, most studies to date have focused on molecular clones with ideal target site recognition and do not account for target site variability observed within and between patients. For clinical success and broad applicability, guide RNA (gRNA) selection must account for circulating strain diversity and incorporate the within-host diversity of HIV.
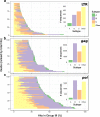
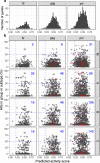
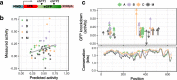
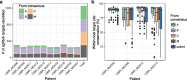
Results: We identified a set of gRNAs targeting HIV LTR, gag, and pol using publicly available sequences for these genes and ranked gRNAs according to global conservation across HIV-1 group M and within subtypes A-C. By considering paired and triplet combinations of gRNAs, we found triplet sets of target sites such that at least one of the gRNAs in the set was present in over 98% of all globally available sequences. We then selected 59 gRNAs from our list of highly conserved LTR target sites and evaluated in vitro activity using a loss-of-function LTR-GFP fusion reporter. We achieved efficient GFP knockdown with multiple gRNAs and found clustering of highly active gRNA target sites near the middle of the LTR. Using published deep-sequence data from HIV-infected patients, we found that globally conserved sites also had greater within-host target conservation. Lastly, we developed a mathematical model based on varying distributions of within-host HIV sequence diversity and enzyme efficacy. We used the model to estimate the number of doses required to deplete the latent reservoir and achieve functional cure thresholds. Our modeling results highlight the importance of within-host target site conservation. While increased doses may overcome low target cleavage efficiency, inadequate targeting of rare strains is predicted to lead to rebound upon cART cessation even with many doses.
Conclusions: Target site selection must account for global and within host viral genetic diversity. Globally conserved target sites are good starting points for design, but multiplexing is essential for depleting quasispecies and preventing viral load rebound upon therapy cessation.
Keywords: CRISPR/Cas9; Computational biology; Endonucleases; Gene editing; Gene therapy; Genomics; HIV; Latent reservoir; Mathematical modeling; Viral genetic diversity.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Gene Therapy with CRISPR/Cas9 Coming to Age for HIV Cure.AIDS Rev. 2017 Oct-Dec;19(3):167-172. AIDS Rev. 2017. PMID: 29019352
-
Novel gRNA design pipeline to develop broad-spectrum CRISPR/Cas9 gRNAs for safe targeting of the HIV-1 quasispecies in patients.Sci Rep. 2019 Nov 19;9(1):17088. doi: 10.1038/s41598-019-52353-9. Sci Rep. 2019. PMID: 31745112 Free PMC article.
-
Broad-Spectrum and Personalized Guide RNAs for CRISPR/Cas9 HIV-1 Therapeutics.AIDS Res Hum Retroviruses. 2018 Nov;34(11):950-960. doi: 10.1089/AID.2017.0274. Epub 2018 Aug 27. AIDS Res Hum Retroviruses. 2018. PMID: 29968495 Free PMC article.
-
CRISPR-CAS Replacing Antiviral Drugs against HIV: An Update.Crit Rev Eukaryot Gene Expr. 2020;30(1):77-83. doi: 10.1615/CritRevEukaryotGeneExpr.2020028233. Crit Rev Eukaryot Gene Expr. 2020. PMID: 32421986 Review.
-
Pathways Toward a Functional HIV-1 Cure: Balancing Promise and Perils of CRISPR Therapy.Methods Mol Biol. 2022;2407:429-445. doi: 10.1007/978-1-0716-1871-4_27. Methods Mol Biol. 2022. PMID: 34985679 Free PMC article. Review.
Cited by
-
Computational analysis of cas proteins unlocks new potential in HIV-1 targeted gene therapy.Front Genome Ed. 2024 Jan 4;5:1248982. doi: 10.3389/fgeed.2023.1248982. eCollection 2023. Front Genome Ed. 2024. PMID: 38239625 Free PMC article.
-
CRISPR/Cas9 Genome Editing to Disable the Latent HIV-1 Provirus.Front Microbiol. 2018 Dec 14;9:3107. doi: 10.3389/fmicb.2018.03107. eCollection 2018. Front Microbiol. 2018. PMID: 30619186 Free PMC article. Review.
-
Impact of misclassified defective proviruses on HIV reservoir measurements.Nat Commun. 2023 Jul 13;14(1):4186. doi: 10.1038/s41467-023-39837-z. Nat Commun. 2023. PMID: 37443365 Free PMC article.
-
Inactivation of Latent HIV-1 Proviral DNA Using Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Treatment and the Assessment of Off-Target Effects.Front Microbiol. 2021 May 26;12:629153. doi: 10.3389/fmicb.2021.629153. eCollection 2021. Front Microbiol. 2021. PMID: 34122355 Free PMC article.
-
Challenges and Promise of Human Immunodeficiency Virus Remission.J Infect Dis. 2021 Feb 15;223(12 Suppl 2):4-12. doi: 10.1093/infdis/jiaa568. J Infect Dis. 2021. PMID: 33586773 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

