Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection
- PMID: 29997209
- PMCID: PMC6146804
- DOI: 10.1128/JVI.00811-18
Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection
Erratum in
-
Correction for Ojwach et al., "Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection".J Virol. 2019 Jan 17;93(3):e01924-18. doi: 10.1128/JVI.01924-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30655384 Free PMC article. No abstract available.
Abstract
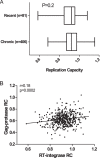
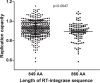
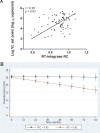
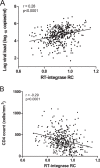
CD8+ T cell-mediated escape mutations in Gag can reduce HIV-1 replication capacity (RC) and alter disease progression, but less is known about immune-mediated attenuation in other HIV-1 proteins. We generated 487 recombinant viruses encoding RT-integrase from individuals with chronic (n = 406) and recent (n = 81) HIV-1 subtype C infection and measured their in vitro RC using a green fluorescent protein (GFP) reporter T cell assay. In recently infected individuals, reverse transcriptase (RT)-integrase-driven RC correlated significantly with viral load set point (r = 0.25; P = 0.03) and CD4+ T cell decline (P = 0.013). Moreover, significant associations between RT integrase-driven RC and viral load (r = 0.28; P < 0.0001) and CD4+ T cell count (r = -0.29; P < 0.0001) remained in chronic infection. In early HIV infection, host expression of the protective HLA-B*81 allele was associated with lower RC (P = 0.05), as was expression of HLA-B*07 (P = 0.02), suggesting early immune-driven attenuation of RT-integrase by these alleles. In chronic infection, HLA-A*30:09 (in linkage disequilibrium with HLA-B*81) was significantly associated with lower RC (P = 0.05), and all 6 HLA-B alleles with the lowest RC measurements represented protective alleles, consistent with long-term effects of host immune pressures on lowering RT-integrase RC. The polymorphisms V241I, I257V, P272K, and E297K in reverse transcriptase and I201V in integrase, all relatively uncommon polymorphisms occurring in or adjacent to optimally described HLA-restricted cytotoxic T-lymphocyte epitopes, were associated with reduced RC. Together, our data suggest that RT-integrase-driven RC is clinically relevant and provide evidence that immune-driven selection of mutations in RT-integrase can compromise RC.IMPORTANCE Identification of viral mutations that compromise HIV's ability to replicate may aid rational vaccine design. However, while certain escape mutations in Gag have been shown to reduce HIV replication and influence clinical progression, less is known about the consequences of mutations that naturally arise in other HIV proteins. Pol is a highly conserved protein, but the impact of Pol function on HIV disease progression is not well defined. Here, we generated recombinant viruses using the RT-integrase region of Pol derived from HIV-1C-infected individuals with recent and chronic infection and measured their ability to replicate in vitro We demonstrate that RT-integrase-driven replication ability significantly impacts HIV disease progression. We further show evidence of immune-mediated attenuation in RT-integrase and identify specific polymorphisms in RT-integrase that significantly decrease HIV-1 replication ability, suggesting which Pol epitopes could be explored in vaccine development.
Keywords: HIV-1 subtype C; HLA polymorphisms; replication capacity.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection.J Virol. 2010 Nov;84(22):11937-49. doi: 10.1128/JVI.01086-10. Epub 2010 Sep 1. J Virol. 2010. PMID: 20810731 Free PMC article.
-
Lack of a significant impact of Gag-Protease-mediated HIV-1 replication capacity on clinical parameters in treatment-naive Japanese individuals.Retrovirology. 2015 Nov 19;12:98. doi: 10.1186/s12977-015-0223-z. Retrovirology. 2015. PMID: 26585907 Free PMC article.
-
HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703.J Immunol. 2011 May 15;186(10):5675-86. doi: 10.4049/jimmunol.1003711. Epub 2011 Apr 15. J Immunol. 2011. PMID: 21498667 Free PMC article.
-
A systematic review of T cell epitopes defined from the proteome of human immunodeficiency virus.Virus Res. 2025 Aug;358:199602. doi: 10.1016/j.virusres.2025.199602. Epub 2025 Jun 23. Virus Res. 2025. PMID: 40562176 Free PMC article. Review.
-
The influence of HLA/HIV genetics on the occurrence of elite controllers and a need for therapeutics geotargeting view.Braz J Infect Dis. 2021 Sep-Oct;25(5):101619. doi: 10.1016/j.bjid.2021.101619. Epub 2021 Sep 22. Braz J Infect Dis. 2021. PMID: 34562387 Free PMC article. Review.
Cited by
-
Subtype-specific differences in Gag-protease replication capacity of HIV-1 isolates from East and West Africa.Retrovirology. 2021 May 5;18(1):11. doi: 10.1186/s12977-021-00554-4. Retrovirology. 2021. PMID: 33952315 Free PMC article.
-
Deep sequencing of the HIV-1 polymerase gene for characterisation of cytotoxic T-lymphocyte epitopes during early and chronic disease stages.Virol J. 2022 Mar 28;19(1):56. doi: 10.1186/s12985-022-01772-8. Virol J. 2022. PMID: 35346259 Free PMC article.
-
First report of computational protein-ligand docking to evaluate susceptibility to HIV integrase inhibitors in HIV-infected Iranian patients.Biochem Biophys Rep. 2022 Mar 29;30:101254. doi: 10.1016/j.bbrep.2022.101254. eCollection 2022 Jul. Biochem Biophys Rep. 2022. PMID: 35368742 Free PMC article.
-
HIV controllers: hope for a functional cure.Front Immunol. 2025 Feb 25;16:1540932. doi: 10.3389/fimmu.2025.1540932. eCollection 2025. Front Immunol. 2025. PMID: 40070826 Free PMC article. Review.
-
Number of HIV-1 founder variants is determined by the recency of the source partner infection.Science. 2020 Jul 3;369(6499):103-108. doi: 10.1126/science.aba5443. Science. 2020. PMID: 32631894 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

