Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells
- PMID: 29997211
- PMCID: PMC6146805
- DOI: 10.1128/JVI.00648-18
Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells
Abstract
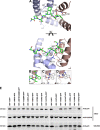
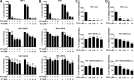
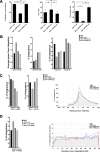
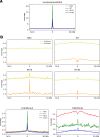
Human immunodeficiency virus type 1 (HIV-1) displays the unique ability to infect nondividing cells. The capsid of HIV-1 is the viral determinant for viral nuclear import. To understand the cellular factors involved in the ability of HIV-1 to infect nondividing cells, we sought to find capsid mutations that allow the virus to infect dividing but not nondividing cells. Because the interaction of capsid with the nucleoporin protein 153 (Nup153) is important for nuclear import of HIV-1, we solved new crystal structures of hexameric HIV-1 capsid in complex with a Nup153-derived peptide containing a phenylalanine-glycine repeat (FG repeat), which we used to guide structure-based mutagenesis of the capsid-binding interface. HIV-1 viruses with mutations in these capsid residues were tested for their ability to infect dividing and nondividing cells. HIV-1 viruses with capsid N57 substitutions infected dividing but not nondividing cells. Interestingly, HIV-1 viruses with N57 mutations underwent reverse transcription but not nuclear translocation. The mutant capsids also lost the ability to interact with Nup153 and CPSF6. The use of small molecules PF74 and BI-2 prevented the interaction of FG-containing nucleoporins (Nups), such as Nup153, with the HIV-1 core. Analysis of integration sites in HIV-1 viruses with N57 mutations revealed diminished integration into transcriptionally active genes in a manner resembling that of HIV-1 in CPSF6 knockout cells or that of HIV-1-N74D. The integration pattern of the N57 mutant HIV-1 can be explained by loss of capsid interaction with CPSF6, whereas capsid interaction with Nup153 is required for HIV-1 to infect nondividing cells. Additionally, the observed viral integration profiles suggested that integration site selection is a multiparameter process that depends upon nuclear factors and the state of the cellular chromatin.IMPORTANCE One of the key advantages that distinguish lentiviruses, such as HIV-1, from all other retroviruses is its ability to infect nondividing cells. Interaction of the HIV-1 capsid with Nup153 and CPSF6 is important for nuclear entry and integration; however, the contribution of each of these proteins to nuclear import and integration is not clear. Using genetics, we demonstrated that these proteins contribute to different processes: Nup153 is essential for the HIV-1 nuclear import in nondividing cells, and CPSF6 is important for HIV-1 integration. In addition, nuclear factors such as CPSF6 and the state of the chromatin are known to be important for integration site selection; nevertheless, the preferential determinant influencing integration site selection is not known. This work demonstrates that integration site selection is a multiparameter process that depends upon nuclear factors and the state of the cellular chromatin.
Keywords: CPSF6; HIV integration; HIV nuclear import; HIV-1; NPC; Nup153; capsid binding; integration; nondividing cells; nuclear import.
Copyright © 2018 American Society for Microbiology.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

