Multiple Layers of CDK5R1 Regulation in Alzheimer's Disease Implicate Long Non-Coding RNAs
- PMID: 29997370
- PMCID: PMC6073344
- DOI: 10.3390/ijms19072022
Multiple Layers of CDK5R1 Regulation in Alzheimer's Disease Implicate Long Non-Coding RNAs
Abstract
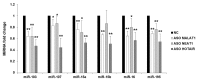
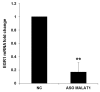
Cyclin-dependent kinase 5 regulatory subunit 1 (CDK5R1) gene encodes for p35, the main activator of Cyclin-dependent kinase 5 (CDK5). The active p35/CDK5 complex is involved in numerous aspects of brain development and function, and its deregulation is closely associated to Alzheimer's disease (AD) onset and progression. We recently showed that miR-15/107 family can negatively regulate CDK5R1 expression modifying mRNA stability. Interestingly, miRNAs belonging to miR-15/107 family are downregulated in AD brain while CDK5R1 is upregulated. Long non-coding RNAs (lncRNAs) are emerging as master regulators of gene expression, including miRNAs, and their dysregulation has been implicated in the pathogenesis of AD. Here, we evaluated the existence of an additional layer of CDK5R1 expression regulation provided by lncRNAs. In particular, we focused on three lncRNAs potentially regulating CDK5R1 expression levels, based on existing data: NEAT1, HOTAIR, and MALAT1. We demonstrated that NEAT1 and HOTAIR negatively regulate CDK5R1 mRNA levels, while MALAT1 has a positive effect. We also showed that all three lncRNAs positively control miR-15/107 family of miRNAs. Moreover, we evaluated the expression of NEAT1, HOTAIR, and MALAT1 in AD and control brain tissues. Interestingly, NEAT1 displayed increased expression levels in temporal cortex and hippocampus of AD patients. Interestingly, we observed a strong positive correlation between CDK5R1 and NEAT1 expression levels in brain tissues, suggesting a possible neuroprotective role of NEAT1 in AD to compensate for increased CDK5R1 levels. Overall, our work provides evidence of another level of CDK5R1 expression regulation mediated by lncRNAs and points to NEAT1 as a biomarker, as well as a potential pharmacological target for AD therapy.
Keywords: Alzheimer’s disease; CDK5R1; HOTAIR; MALAT1; NEAT1; lncRNAs; miR-15/107.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

