Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China
- PMID: 29998087
- PMCID: PMC6030573
- DOI: 10.3389/fcimb.2018.00219
Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China
Abstract
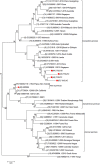
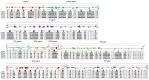
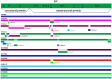
In 2015, a dengue outbreak with 1,067 reported cases occurred in Xishuangbanna, a city in China that borders Burma and Laos. To characterize the virus, the complete genome sequence was obtained and phylogenetic, mutation, substitution and recombinant analyses were performed. DENV-NS1 positive serum samples were collected from dengue fever patients, and complete genome sequences were obtained through RT-qPCR from these serum samples. Phylogenetic trees were then constructed by maximum likelihood phylogeny test (MEGA7.0), followed by analysis of nucleotide mutation and amino acid substitution. The recombination events among DENVs were also analyzed by RDP4 package. The diversity analysis of secondary structure for translated viral proteins was also performed. The complete genome sequences of four amplified viruses (YNXJ10, YNXJ12, YNXJ13, and YNXJ16) were 10,742, 10,742, 10,741, and 10,734 nucleotides in length, and phylogenetic analysis classified the viruses as cosmopolitan genotype of DENV-2. All viruses were close to DENV Singapore 2013 (KX380828.1) and the DENV China 2013 (KF479233.1). In comparison to DENV-2SS (M29095), the total numbers of base substitutions were 712 nt (YNXJ10), 809 nt (YNXJ12), 772 nt (YNXJ13), and 841 nt (YNXJ16), resulting in 109, 171, 130, and 180 amino acid substitutions in translated regions, respectively. In addition, compared with KX380828.1, there were 44, 105, 64, and 116 amino acid substitutions in translated regions, respectively. The highest mutation rate occurred in the prM region, and the lowest mutation rate occurred in the NS4B region. Most of the recombination events occurred in the prM, E and NS2B/3 regions, which corresponded with the mutation frequency of the related portion. Secondary structure prediction within the 3,391 amino acids of DENV structural proteins showed there were 7 new possible nucleotide-binding sites and 6 lost sites compared to DENV-2SS. In addition, 41 distinct amino acid changes were found in the helix regions, although the distribution of the exposed and buried regions changed only slightly. Our findings may help to understand the intrinsic geographical relatedness of DENV-2 and contributes to the understanding of viral evolution and its impact on the epidemic potential and pathogenicity of DENV.
Keywords: characterization; dengue virus; genome; phylogenetic analysis; recombinant analysis.
Figures


Similar articles
-
Complete genome analysis of dengue virus type 3 isolated from the 2013 dengue outbreak in Yunnan, China.Virus Res. 2017 Jun 15;238:164-170. doi: 10.1016/j.virusres.2017.06.015. Epub 2017 Jun 23. Virus Res. 2017. PMID: 28648850
-
Complete Genome Characterization of the 2017 Dengue Outbreak in Xishuangbanna, a Border City of China, Burma and Laos.Front Cell Infect Microbiol. 2018 May 8;8:148. doi: 10.3389/fcimb.2018.00148. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29868504 Free PMC article.
-
Molecular characterization of the viral structural protein genes in the first outbreak of dengue virus type 2 in Hunan Province, inland China in 2018.BMC Infect Dis. 2021 Feb 10;21(1):166. doi: 10.1186/s12879-021-05823-3. BMC Infect Dis. 2021. PMID: 33568111 Free PMC article.
-
Molecular characterization of an imported dengue virus serotype 4 isolate from Thailand.Arch Virol. 2018 Oct;163(10):2903-2906. doi: 10.1007/s00705-018-3906-7. Epub 2018 Jun 14. Arch Virol. 2018. PMID: 29948381 Review.
-
Unraveling Dengue Virus Diversity in Asia: An Epidemiological Study through Genetic Sequences and Phylogenetic Analysis.Viruses. 2024 Jun 28;16(7):1046. doi: 10.3390/v16071046. Viruses. 2024. PMID: 39066210 Free PMC article. Review.
Cited by
-
Detection of a Multiple Circulation Event of Dengue Virus 2 Strains in the Northern Region of Brazil.Trop Med Infect Dis. 2024 Jan 9;9(1):17. doi: 10.3390/tropicalmed9010017. Trop Med Infect Dis. 2024. PMID: 38251214 Free PMC article.
-
Genomic characterization of dengue virus serotype 2 during dengue outbreak and endemics in Hangzhou, Zhejiang (2017-2019).Front Microbiol. 2023 Aug 25;14:1245416. doi: 10.3389/fmicb.2023.1245416. eCollection 2023. Front Microbiol. 2023. PMID: 37692383 Free PMC article.
-
Isolation and molecular characterization of dengue virus clinical isolates from pediatric patients in New Delhi.Int J Infect Dis. 2019 Jul;84S:S25-S33. doi: 10.1016/j.ijid.2018.12.003. Epub 2018 Dec 7. Int J Infect Dis. 2019. PMID: 30528666 Free PMC article.
-
Seroepidemiologic study on convalescent sera from dengue fever patients in Jinghong, Yunnan.Virol Sin. 2022 Feb;37(1):19-29. doi: 10.1016/j.virs.2021.12.001. Epub 2021 Dec 17. Virol Sin. 2022. PMID: 35234619 Free PMC article.
-
Genomic variants and molecular epidemiological characteristics of dengue virus in China revealed by genome-wide analysis.Virus Evol. 2025 Mar 17;11(1):veaf013. doi: 10.1093/ve/veaf013. eCollection 2025. Virus Evol. 2025. PMID: 40135062 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

