High-Throughput Metabolomics by 1D NMR
- PMID: 29999221
- PMCID: PMC6391965
- DOI: 10.1002/anie.201804736
High-Throughput Metabolomics by 1D NMR
Abstract
Metabolomics deals with the whole ensemble of metabolites (the metabolome). As one of the -omic sciences, it relates to biology, physiology, pathology and medicine; but metabolites are chemical entities, small organic molecules or inorganic ions. Therefore, their proper identification and quantitation in complex biological matrices requires a solid chemical ground. With respect to for example, DNA, metabolites are much more prone to oxidation or enzymatic degradation: we can reconstruct large parts of a mammoth's genome from a small specimen, but we are unable to do the same with its metabolome, which was probably largely degraded a few hours after the animal's death. Thus, we need standard operating procedures, good chemical skills in sample preparation for storage and subsequent analysis, accurate analytical procedures, a broad knowledge of chemometrics and advanced statistical tools, and a good knowledge of at least one of the two metabolomic techniques, MS or NMR. All these skills are traditionally cultivated by chemists. Here we focus on metabolomics from the chemical standpoint and restrict ourselves to NMR. From the analytical point of view, NMR has pros and cons but does provide a peculiar holistic perspective that may speak for its future adoption as a population-wide health screening technique.
Keywords: NMR; fingerprinting; metabolomics; omic sciences; profiling.
© 2018 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
The authors declare no conflict of interest.
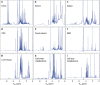
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

