A bidirectional relationship between sleep and oxidative stress in Drosophila
- PMID: 30001323
- PMCID: PMC6042693
- DOI: 10.1371/journal.pbio.2005206
A bidirectional relationship between sleep and oxidative stress in Drosophila
Abstract
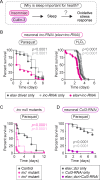
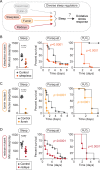
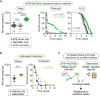
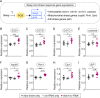
Although sleep appears to be broadly conserved in animals, the physiological functions of sleep remain unclear. In this study, we sought to identify a physiological defect common to a diverse group of short-sleeping Drosophila mutants, which might provide insight into the function and regulation of sleep. We found that these short-sleeping mutants share a common phenotype of sensitivity to acute oxidative stress, exhibiting shorter survival times than controls. We further showed that increasing sleep in wild-type flies using genetic or pharmacological approaches increases survival after oxidative challenge. Moreover, reducing oxidative stress in the neurons of wild-type flies by overexpression of antioxidant genes reduces the amount of sleep. Together, these results support the hypothesis that a key function of sleep is to defend against oxidative stress and also point to a reciprocal role for reactive oxygen species (ROS) in neurons in the regulation of sleep.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

