Stable-Isotope Probing of Human and Animal Microbiome Function
- PMID: 30001854
- PMCID: PMC6249988
- DOI: 10.1016/j.tim.2018.06.004
Stable-Isotope Probing of Human and Animal Microbiome Function
Abstract
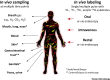
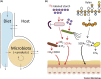
Humans and animals host diverse communities of microorganisms important to their physiology and health. Despite extensive sequencing-based characterization of host-associated microbiomes, there remains a dramatic lack of understanding of microbial functions. Stable-isotope probing (SIP) is a powerful strategy to elucidate the ecophysiology of microorganisms in complex host-associated microbiotas. Here, we suggest that SIP methodologies should be more frequently exploited as part of a holistic functional microbiomics approach. We provide examples of how SIP has been used to study host-associated microbes in vivo and ex vivo. We highlight recent developments in SIP technologies and discuss future directions that will facilitate deeper insights into the function of human and animal microbiomes.
Keywords: NanoSIMS; RNA-SIP; Raman microspectroscopy; gut microbiota; single-cell imaging.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Hehemann J.-H. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. - PubMed
-
- Turaev D., Rattei T. High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr. Opin. Biotechnol. 2016;39:174–181. - PubMed
-
- Li S.S. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

