Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism
- PMID: 30001904
- PMCID: PMC6109460
- DOI: 10.1016/j.tem.2018.06.002
Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism
Abstract
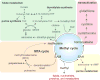
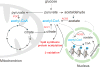
Epigenetic modifications on chromatin are most commonly thought to be involved in the transcriptional regulation of gene expression. Due to their dependency on small-molecule metabolites, these modifications can relay information about cellular metabolic state to the genome for the activation or repression of particular sets of genes. In this review we discuss emerging evidence that these modifications might also have a metabolic purpose. Due to their abundance, the histones have the capacity to store substantial amounts of useful metabolites or to enable important metabolic transformations. Such metabolic functions for histones could help to explain the widespread occurrence of particular modifications that may not always be strongly correlated with transcriptional activity.
Keywords: SAM; acetate; acetyl-CoA; epigenetics; histone acetylation; histone methylation.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

References
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. - PubMed
-
- German DC, Bloch CA, Kredich NM. Measurements of S-adenosylmethionine and L-homocysteine metabolism in cultured human lymphoid cells. J Biol Chem. 1983;258(18):10997–1003. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

