Chromatin Loop Formation Induced by a Subtelomeric Protosilencer Represses EPA Genes in Candida glabrata
- PMID: 30002080
- PMCID: PMC6116953
- DOI: 10.1534/genetics.118.301202
Chromatin Loop Formation Induced by a Subtelomeric Protosilencer Represses EPA Genes in Candida glabrata
Abstract
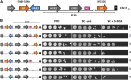
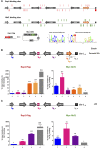
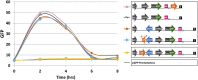
Adherence, an important virulence factor, is mediated by the EPA (Epithelial Adhesin) genes in the opportunistic pathogen Candida glabrata Expression of adhesin-encoding genes requires tight regulation to respond to harsh environmental conditions within the host. The majority of EPA genes are localized in subtelomeric regions regulated by subtelomeric silencing, which depends mainly on Rap1 and the Sir proteins. In vitro adhesion to epithelial cells is primarily mediated by Epa1. EPA1 forms a cluster with EPA2 and EPA3 in the right telomere of chromosome E (E-R). This telomere contains a cis-acting regulatory element, the protosilencer Sil2126 between EPA3 and the telomere. Interestingly, Sil2126 is only active in the context of its native telomere. Replacement of the intergenic regions between EPA genes in E-R revealed that cis-acting elements between EPA2 and EPA3 are required for Sil2126 activity when placed 32 kb away from the telomere (Sil@-32kb). Sil2126 contains several putative binding sites for Rap1 and Abf1, and its activity depends on these proteins. Indeed, Sil2126 binds Rap1 and Abf1 at its native position and also when inserted at -32 kb, a silencing-free environment in the parental strain. In addition, we found that Sil@-32kb and Sil2126 at its native position can physically interact with the intergenic regions between EPA1-EPA2 and EPA2-EPA3 respectively, by chromosome conformation capture assays. We speculate that Rap1 and Abf1 bound to Sil2126 can recruit the Silent Information Regulator complex, and together mediate silencing in this region, probably through the formation of a chromatin loop.
Keywords: Candida glabrata; EPA genes; Rap1; chromatin loop; cis-elements; protosilencer; transcriptional regulation.
Copyright © 2018 by the Genetics Society of America.
Figures






References
-
- Ausubel, F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G. et al. , (Editors) 2001. Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken, NJ.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

