Mitotic Gene Bookmarking: An Epigenetic Program to Maintain Normal and Cancer Phenotypes
- PMID: 30002192
- PMCID: PMC6214712
- DOI: 10.1158/1541-7786.MCR-18-0415
Mitotic Gene Bookmarking: An Epigenetic Program to Maintain Normal and Cancer Phenotypes
Abstract
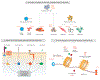
Reconfiguration of nuclear structure and function during mitosis presents a significant challenge to resume the next cell cycle in the progeny cells without compromising structural and functional identity of the cells. Equally important is the requirement for cancer cells to retain the transformed phenotype, that is, unrestricted proliferative potential, suppression of cell phenotype, and activation of oncogenic pathways. Mitotic gene bookmarking retention of key regulatory proteins that include sequence-specific transcription factors, chromatin-modifying factors, and components of RNA Pol (RNAP) I and II regulatory machineries at gene loci on mitotic chromosomes plays key roles in coordinate control of cell phenotype, growth, and proliferation postmitotically. There is growing recognition that three distinct protein types, mechanistically, play obligatory roles in mitotic gene bookmarking: (i) Retention of phenotypic transcription factors on mitotic chromosomes is essential to sustain lineage commitment; (ii) Select chromatin modifiers and posttranslational histone modifications/variants retain competency of mitotic chromatin for gene reactivation as cells exit mitosis; and (iii) Functional components of RNAP I and II transcription complexes (e.g., UBF and TBP, respectively) are retained on genes poised for reactivation immediately following mitosis. Importantly, recent findings have identified oncogenes that are associated with target genes on mitotic chromosomes in cancer cells. The current review proposes that mitotic gene bookmarking is an extensively utilized epigenetic mechanism for stringent control of proliferation and identity in normal cells and hypothesizes that bookmarking plays a pivotal role in maintenance of tumor phenotypes, that is, unrestricted proliferation and compromised control of differentiation. Mol Cancer Res; 16(11); 1617-24. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

