Proteomic Analysis of NCK1/2 Adaptors Uncovers Paralog-specific Interactions That Reveal a New Role for NCK2 in Cell Abscission During Cytokinesis
- PMID: 30002203
- PMCID: PMC6166670
- DOI: 10.1074/mcp.RA118.000689
Proteomic Analysis of NCK1/2 Adaptors Uncovers Paralog-specific Interactions That Reveal a New Role for NCK2 in Cell Abscission During Cytokinesis
Abstract
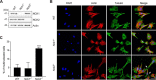
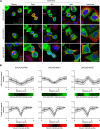
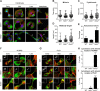
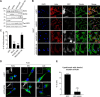
Signals from cell surface receptors are often relayed via adaptor proteins. NCK1 and NCK2 are Src-Homology (SH) 2 and 3 domain adaptors that regulate processes requiring a remodeling of the actin cytoskeleton. Evidence from gene inactivation in mouse suggests that NCK1 and NCK2 are functionally redundant, although recent reports support the idea of unique functions for NCK1 and NCK2. We sought to examine this question further by delineating NCK1- and NCK2-specific signaling networks. We used both affinity purification-mass spectrometry and BioID proximity labeling to identify NCK1/2 signaling networks comprised of 98 proteins. Strikingly, we found 30 proteins restricted to NCK1 and 28 proteins specifically associated with NCK2, suggesting differences in their function. We report that Nck2-/-, but not Nck1-/- mouse embryo fibroblasts (MEFs) are multinucleated and display extended protrusions reminiscent of intercellular bridges, which correlate with an extended time spent in cytokinesis as well as a failure of a significant proportion of cells to complete abscission. Our data also show that the midbody of NCK2-deficient cells is not only increased in length, but also altered in composition, as judged by the mislocalization of AURKB, PLK1 and ECT2. Finally, we show that NCK2 function during cytokinesis requires its SH2 domain. Taken together, our data delineate the first high-confidence interactome for NCK1/2 adaptors and highlight several proteins specifically associated with either protein. Thus, contrary to what is generally accepted, we demonstrate that NCK1 and NCK2 are not completely redundant, and shed light on a previously uncharacterized function for the NCK2 adaptor protein in cell division.
Keywords: Affinity proteomics; Cell biology; Cell division; Imaging; Mass Spectrometry; Networks; Phosphorylation; Protein-Protein Interactions; SH2/SH3 Domains; Signal Transduction.
© 2018 Jacquet et al.
Figures

References
-
- Buday L., Wunderlich L., and Tamas P. (2002) The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal 14, 723–731 - PubMed
-
- Frese S., Schubert W. D., Findeis A. C., Marquardt T., Roske Y. S., Stradal T. E., and Heinz D. W. (2006) The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236–18245 - PubMed
-
- Chen M., She H., Davis E. M., Spicer C. M., Kim L., Ren R., Le Beau M. M., and Li W. (1998) Identification of Nck family genes, chromosomal localization, expression, and signaling specificity. J. Biol. Chem. 273, 25171–25178 - PubMed
-
- Li W., Fan J., and Woodley D. T. (2001) Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene 20, 6403–6417 - PubMed
-
- Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., and Mayer B. J. (2001) Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276, 26448–26452 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

