Targeted Approaches for In Situ Gut Microbiome Manipulation
- PMID: 30002345
- PMCID: PMC6071227
- DOI: 10.3390/genes9070351
Targeted Approaches for In Situ Gut Microbiome Manipulation
Abstract
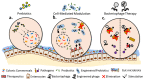
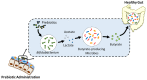
Microbial communities and their collective genomes form the gut microbiome, of which bacteria are the major contributor. Through their secreted metabolites, bacteria interact with the host, influencing human health and physiology. Perturbation of the microbiota and metabolome has been associated with various diseases and metabolic conditions. As knowledge on fundamental host-microbiome interactions and genetic engineering tools becomes readily available, targeted manipulation of the gut microbiome for therapeutic applications gains favourable attention. Manipulation of the gut microbiome can be achieved by altering the microbiota population and composition, or by modifying the functional metabolic activity of the microbiome to promote health and restore the microbiome balance. In this article, we review current works that demonstrate various strategies employed to manipulate the gut microbiome in situ to various degrees of precision.
Keywords: CRISPR-Cas9; gut microbiome; microbiome modulation; phage; prebiotics; probiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bielecka M. Probiotics in Food. In: Sikorski Z., editor. Chemical and Functional Properties of Food Components. 3rd ed. CRC Press; Boca Raton, FL, USA: 2007. pp. 413–426. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

