L1 retrotransposition in the soma: a field jumping ahead
- PMID: 30002735
- PMCID: PMC6035798
- DOI: 10.1186/s13100-018-0128-1
L1 retrotransposition in the soma: a field jumping ahead
Abstract
Retrotransposons are transposable elements (TEs) capable of "jumping" in germ, embryonic and tumor cells and, as is now clearly established, in the neuronal lineage. Mosaic TE insertions form part of a broader landscape of somatic genome variation and hold significant potential to generate phenotypic diversity, in the brain and elsewhere. At present, the LINE-1 (L1) retrotransposon family appears to be the most active autonomous TE in most mammals, based on experimental data obtained from disease-causing L1 mutations, engineered L1 reporter systems tested in cultured cells and transgenic rodents, and single-cell genomic analyses. However, the biological consequences of almost all somatic L1 insertions identified thus far remain unknown. In this review, we briefly summarize the current state-of-the-art in the field, including estimates of L1 retrotransposition rate in neurons. We bring forward the hypothesis that an extensive subset of retrotransposition-competent L1s may be de-repressed and mobile in the soma but largely inactive in the germline. We discuss recent reports of non-canonical L1-associated sequence variants in the brain and propose that the elevated L1 DNA content reported in several neurological disorders may predominantly comprise accumulated, unintegrated L1 nucleic acids, rather than somatic L1 insertions. Finally, we consider the main objectives and obstacles going forward in elucidating the biological impact of somatic retrotransposition.
Keywords: Genomics; L1; LINE-1; Mosaicism; Neurobiology; Retrotransposon.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
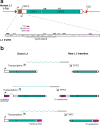
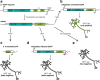

References
-
- Belzile F, Yoder JI. Pattern of somatic transposition in a high copy ac tomato line. Plant J. 1992;2:173–179. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

