Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases
- PMID: 30002876
- PMCID: PMC6037748
- DOI: 10.1038/s41525-018-0053-8
Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases
Abstract
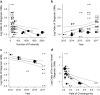
Genetic diseases are leading causes of childhood mortality. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are relatively new methods for diagnosing genetic diseases, whereas chromosomal microarray (CMA) is well established. Here we compared the diagnostic utility (rate of causative, pathogenic, or likely pathogenic genotypes in known disease genes) and clinical utility (proportion in whom medical or surgical management was changed by diagnosis) of WGS, WES, and CMA in children with suspected genetic diseases by systematic review of the literature (January 2011-August 2017) and meta-analysis, following MOOSE/PRISMA guidelines. In 37 studies, comprising 20,068 children, diagnostic utility of WGS (0.41, 95% CI 0.34-0.48, I2 = 44%) and WES (0.36, 95% CI 0.33-0.40, I2 = 83%) were qualitatively greater than CMA (0.10, 95% CI 0.08-0.12, I2 = 81%). Among studies published in 2017, the diagnostic utility of WGS was significantly greater than CMA (P < 0.0001, I2 = 13% and I2 = 40%, respectively). Among studies featuring within-cohort comparisons, the diagnostic utility of WES was significantly greater than CMA (P < 0.001, I2 = 36%). The diagnostic utility of WGS and WES were not significantly different. In studies featuring within-cohort comparisons of WGS/WES, the likelihood of diagnosis was significantly greater for trios than singletons (odds ratio 2.04, 95% CI 1.62-2.56, I2 = 12%; P < 0.0001). Diagnostic utility of WGS/WES with hospital-based interpretation (0.42, 95% CI 0.38-0.45, I2 = 48%) was qualitatively higher than that of reference laboratories (0.29, 95% CI 0.27-0.31, I2 = 49%); this difference was significant among studies published in 2017 (P < .0001, I2 = 22% and I2 = 26%, respectively). The clinical utility of WGS (0.27, 95% CI 0.17-0.40, I2 = 54%) and WES (0.17, 95% CI 0.12-0.24, I2 = 76%) were higher than CMA (0.06, 95% CI 0.05-0.07, I2 = 42%); this difference was significant for WGS vs CMA (P < 0.0001). In conclusion, in children with suspected genetic diseases, the diagnostic and clinical utility of WGS/WES were greater than CMA. Subgroups with higher WGS/WES diagnostic utility were trios and those receiving hospital-based interpretation. WGS/WES should be considered a first-line genomic test for children with suspected genetic diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- March of Dimes. March of Dimes Data Book for Policy Makers: Maternal, Infant, and Child Health in the United States. Office of Government Affairs, March of Dimes (Washington, DC, 2016).
-
- Xu J, Murphy SL, Kochanek KD, Arias E. Mortality in the United States, 2015. NCHS Data Brief. 2016;267:1–8. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

